-
Notifications
You must be signed in to change notification settings - Fork 11
NIRSTORM Mini Course 2020 (PERFORM Center)
NIRSTORM is a Brainstorm plugin for fNIRS data analysis.
- Thomas Vincent, EPIC center, Montreal Heart Institute, Montreal, Canada
- Zhengchen Cai, Physics Department and PERFORM Center, Concordia University, Montreal, Canada
- Edouard Delaire, Physics Department and PERFORM Center, Concordia University, Montreal, Canada
- Amanda Spilkin, Physics Department and PERFORM Center, Concordia University, Montreal, Canada
- Christophe Grova, Physics Department and PERFORM Center, Concordia University, Montreal, Canada
- Hardware: a basic recent laptop should be fine.
- An internet connection. A dedicated wifi connection will be available on site for the participants of the workshop.
- Operating system: Linux, windows or Mac OS. Everything should work on all systems though we recommend using Linux since more tests have been done with it.
- Disk space: the full installation with dependencies and sample data should take no more than 4Gb. Some space can be freed by cleaning the CPLEX dependency (saves ~3Gb).
-
Matlab: version > R2015b is recommended.
The Matlab Curve Fitting Toolbox is required to perform motion correction. If you don't have it, you may skip this step in the first tutorial. The provided data do not contain too much movement. - Before attending the workshop: Please complete the following Setup section.
If Brainstorm is already installed, update to the latest version (Menu bar: Update > Update Brainstorm).
For a new installation, Brainstorm requires a user to be registered in order to download it. This allows their development team to track their usage. You can proceed with the registration at this page.
Then use the newly created account to download the latest version of brainstorm here. On this page under "Software", download the archive named like "brainstorm_180501.zip" (~60Mb).
Uncompress this archive to a folder of your choice on your local drive, eg your Documents folder. Then run matlab and change the current directory of matlab to the one where the archive was uncompressed. This can be done either via the folder selector or by a command (replace <current user> with your actual user name):
-
Windows:
cd C:\Users\<current user>\Documents\ -
Mac OS:
cd /Users/<current user>/Documents/ -
Linux:
cd /home/<current user>/Documents/
Once in the correct brainstorm folder, type the command brainstorm and agree to the license.
For the first time it runs, brainstorm asks to specify a database folder. This will be the place where all the data imported by brainstorm will go. Create a folder named brainstorm_db in eg your Documents folder and specify it as your main brainstorm database. Then, the main brainstorm interface will be displayed.
Note: you can avoid having to change to the brainstorm directory every time. To do so, while brainstorm is running, go to the "Home" tab of Matlab then "Set path". There click on "Save" and "Close". Now you're able to run brainstorm without changing to the installation directory every time.
No prior knowledge of brainstorm is mandatory for the current workshop session which should cover the basics. However, if one wishes to get familiar with brainstorm features, we refer to the brainstorm introduction page.
Important for mac users: If the right-click doesn't work anywhere in the Brainstorm interface and you cannot get to see the popup menus in the database explorer, try to change the configuration of your trackpad in the system preferences and enable left-click. Alternatively, connect a standard external mouse with two buttons. Some Apple pointing devices do not interact very well with Java/Matlab.
CPLEX is a set of tools to perform functional optimization edited by IBM. It is used here to solve the integer programming problem during the optimal montage computation (last tutorial of this workshop).
For license concerns, we cannot bundle the CPLEX library within NIRSTORM. One can purchase a free copy of CPLEX if they are students or academics. To do so, go to the IBM webstore and click on "register" at the top right. It will ask for your institutional information. Then from this page, get the version of CPLEX that corresponds to your status (student or faculty) and system configuration.
Once installed, the location of the CPLEX matlab bindings that are needed for nirstorm depends on your system:
-
On windows:
C:\Program Files\IBM\ILOG\CPLEX_Studio128\cplex\matlab\x64_win64 -
On Mac OS:
Applications/CPLEX_Studio128/cplex/matlab/x86-64_osx -
On linux (from the folder where you installed CPLEX studio):
cplex_studio_128/cplex/matlab/x86-64_linux
Note that the whole CPLEX studio package takes around 700 Mb to download and requires around 3 Gb of drive space for installation. However, if disk space is an issue, one can extract only the Matlab bindings needed by nirstorm (less than 25 Mb). To do so, copy the content of the path specified above into eg your Documents folder. After that, you can uninstall CPLEX studio.
Finally, add the path to the CPLEX Matlab bindings to the Matlab path: go to the "Home" tab, click "Set path" (yellow folder icon) then click on "Add with subfolders" and select the folder of the CPLEX Matlab bindings.
If you cannot get a working version of CPLEX, let us know during the workshop and we'll try to help you.
Go to the release page of the project and download the latest version (eg 0.51). Click on "Source code (zip)" in the Asset section.
Uncompress the archive in the folder of your choice, eg your Documents folder. Then change the Matlab directory to the folder where you uncompressed the archive, eg Documents/nirstorm-0.51. Make sure brainstorm is running and run the following command in the Matlab prompt:
nst_install('copy');
This will install all NIRSTORM processes into Brainstorm.
Note: If you have trouble during this installation step, you can also copy the content of the bst_plugin folder inside your brainstorm folder (eg .brainstrom/process)
All data required for this session are regrouped in an archive of ~500 Mb. The total disk space required after data download and installation is ~900Mb. The size is quite large because of optical data which are required for the computation of optimal montages.
** Licence for Module 1, 3, and 4:** This tutorial dataset (NIRS and MRI data) remains proprietary of the MultiFunkIm Laboratory of Dr Grova, at PERFORM centre, Concordia University, Montreal. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from our group. For questions please contact Christophe Grova: [email protected]
Download Nirstorm_workshop_PERFORM_2020.zip and uncompress it to your Document folder. You should obtain a sub-folder named Nirstorm_workshop_PERFORM_2020.
It contains three motor sample data :
- One for the classic channel-space analysis tutorial
- One for the cortical reconstruction tutorial It also contains :
- a modified version of the Colin27 template (for the optimal montage tutorial)
- optical data specific to the Colin27 template (for the optimal montage tutorial) Once data is extracted, you may delete the archive.
Warning: the unzipping tool you used (especially under windows) may have created an extra directory level like Nirstorm_workshop_PERFORM_2020\Nirstorm_workshop_PERFORM_2020. Make sure to select the deepest folder.
** Licence for Module 2:** This tutorial dataset (NIRS) remains proprietary of the CNRS / CENIR MEG-EEG at ICM, Paris. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from our group. For questions please contact Laurent Hugueville: [email protected]
Module 2 contains one motor sample data used for the GLM channel-space analysis tutorial at the subject and group level
Note: Data for module 2 will be provided later during the course using SLACK or USB key.
Here is a cheat sheet on the main brainstorm features that will be used during the tutorials:
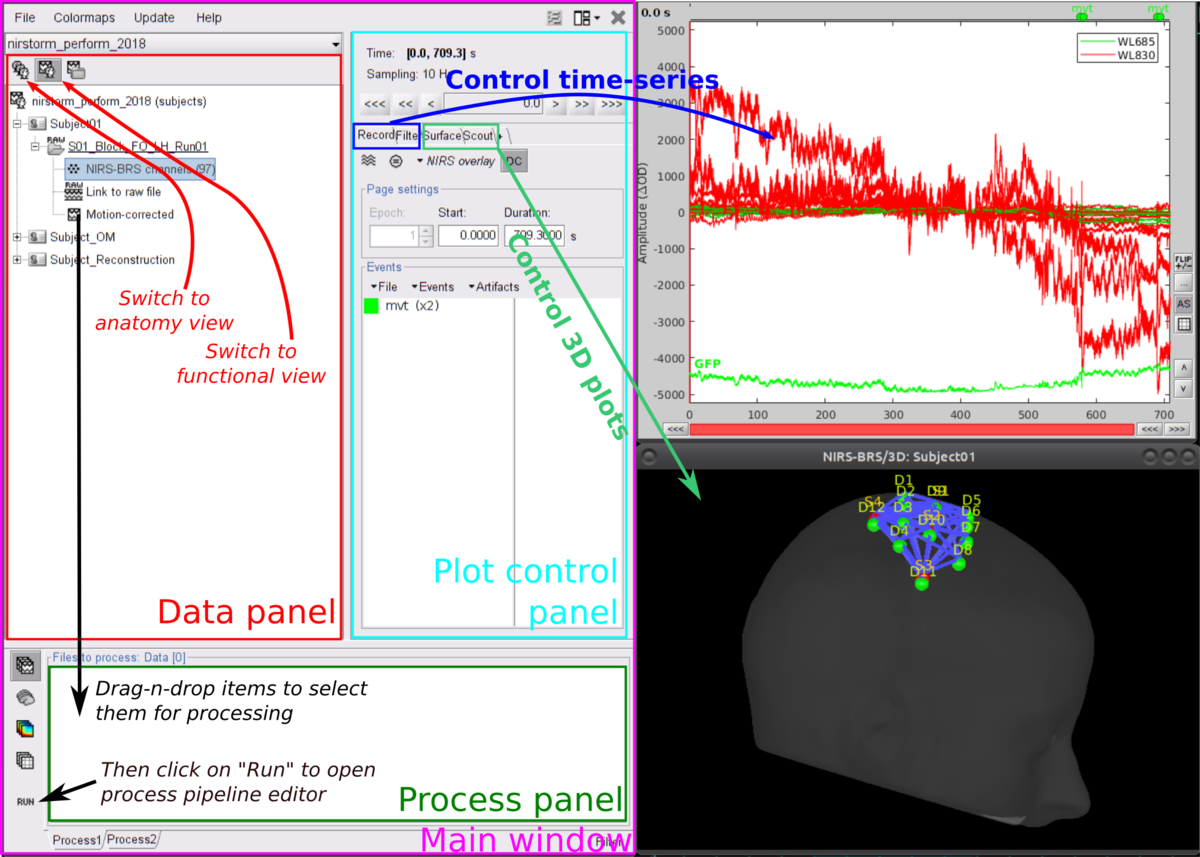
Follow the tutorial on basic NIRS data analysis in the channel space. This link points directly to the data importation section and skips the first parts. You don't need to follow the setup sections of this tutorial since all details should have been covered previously in the current wiki page.
Important: you will find the data set required for this tutorial in the downloaded sample data folder: Nirstorm_workshop_PERFORM_2020/channel_space_analysis/sample_nirs.
Note: if you don't have the Matlab Curve Fitting Toolbox, skip the motion correction step. @todo: ask brainstorm to change color for HbO/HbR
This module uses a grasping task to illustrate how to apply the General Linear Model in NIRSTORM to perform statistical analysis in the channel space. This module will cover both individual analysis and group analysis
- 6 right-handed subject
- 5 minutes of baseline followed by a block-design dominate hand grasping task.
- Block design: 15 to 20 secs of rest followed by 15 secs of task. Repeated 15 times
- 30 channels per wavelength covering the visual area and the two motor cortex [ see figure 1]
- 2 wavelengths: 685nm and 830nm
- Sampling rates: 12.5Hz
This tutorial dataset remains a property of the Spine and Brain Institute (ICM), Paris. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from the ICM Laboratory.
This experiment was conducted in 2017 at ICM by :
- Mélanie Pélégrini-Issac, Sorbonne Université, CNRS, INSERM, Laboratoire d’Imagerie Biomédicale, LIB, F-75006, Paris, France
- Fida Najjar, Sorbonne Université, CNRS, INSERM, Laboratoire d’Imagerie Biomédicale, LIB, F-75006, Paris, France
- Laurent Hugueville, Institut du Cerveau et de la Moelle épinière – ICM, Sorbonne Universités, UPMC Univ Paris 06, Inserm U1127, CNRS UMR 7225, Paris, France
- Denis Schwartz, Institut du Cerveau et de la Moelle épinière – ICM, Sorbonne Universités, UPMC Univ Paris 06, Inserm U1127, CNRS UMR 7225, Paris, France
All the participant signed a written consent form and ethic was approved by the ethics committee of Île-de-France 6 ( CCP number: 14012 )
- From the menu of the Brainstorm main panel: File > Load protocol > Load from zip file > Select the downloaded protocol file: 'Nirstorm_workshop_PERFORM_2020/GLM_analysis/protocol_nirs_glm_subject.zip'
After loading the protocol, your brainstorm database should contain the following files :

- GraspingRight contains the raw data
-
GraspingRight_Hb contains the contains pre-processed data:
- Hb [Topo] contains the variations of concentration of oxy-hemoglobin (HbO), deoxy-hemoglobin (HbR) and total hemoglobin (HbT) from the measured light intensity time courses at different wavelengths. The MBLL is computed using the 5 first minutes of measurement as baseline windows.
- Hb [Topo] | band(0.01-0.1Hz) contains the filtered data filtered between 0.01Hz and 0.1Hz
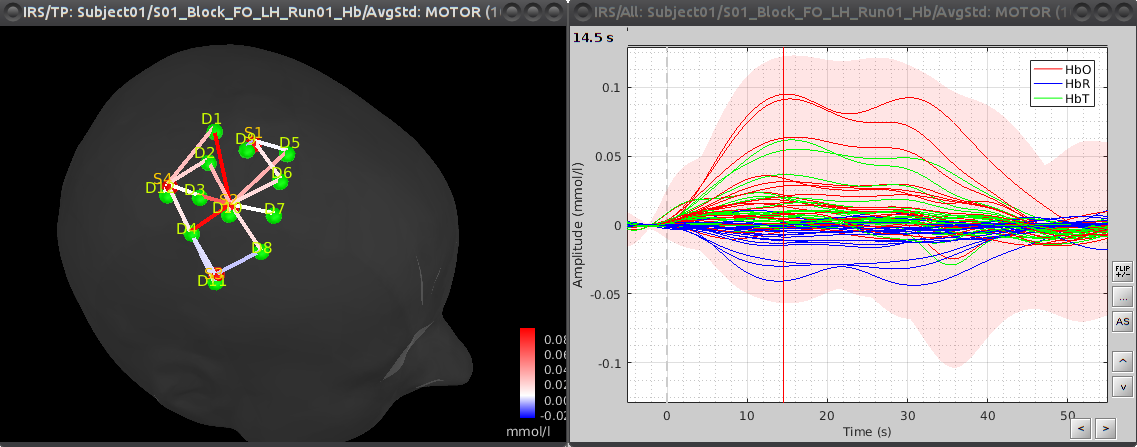
- AvgStd: GraspingRight (14) contains a windows-average of the data centered on each motor task [ see bellow]
To visualize the haemodynamic response, double click on Hb [Topo] | band(0.01-0.1Hz):

The first level analysis relies on the so-called General Linear Model.
The statistical analysis is done in 3 steps :
- First, fitting the GLM model to the data using 'GLM - 1st level design and fit'
- then, compute the contrast using 'GLM - 1st level contrast'
- and finally, compute the Student t-test using 'GLM - contrast t-test'
Drag and drop "Hb [Topo] | band(0.01-0.1Hz)" into the process field and press "Run". In the process menu, select "NIRS > GLM - 1st level design and fit.
Fig. 4 GLM parameters
This process takes the following parameters
-
Signal Information
- Applied High-pass filter: enter the value of the low-cutoff frequency used to remove low-frequency drift ( 0.01 here) Note: This does not reapply a filter to the data
- Ignore starting signal: enter the windows of time that you want to ignore from the beginning of the file. This can be useful if you want to ignore motion at the beginning of your recording or to separate the baseline from the actual task. Here, enter 290s to remove the baseline.
-
Optimization Method
- The GLM is solved using Ordinary Least Square equations.
-
Serial Correlation Pre-processing
- Method: Select the method to handle serial correlation in the noise. Pre-coloring will filter the data with the canonical HRF as define in NIRS-SPM. The pre-whitening method implemented here will remove the noise auto-correlation by iteratively fit an AR(1) model to the noise. Here choose pre-coloring
-
Design Matrix
- Stimulation events: enter the name of the events used to compute the sign matrix. Here enter 'GraspingRight, Rest'
- HRF model: select the model of the haemodynamic response that will be convoluted with your experimental design: Here select canonical
- Add constant regressor: if checked, a constant regressor will be added to the design matrix.
The process will output a file called 'Hb [Topo] | band(0.01-0.1Hz) | GLM OLS_precoloring fitted model'. This file contains all the information about the model that has been fitted: - the B map, the design matrix, the covariance matrix, the residual mse, and the associate degree of freedom that will be used to perform the Student's t-test.
You can also ask the process to store the following additional information :
- Residual: contains the residual data that cannot be explained by the model (eg noise)
- Fit: contains the signal explained by the model (X*B)
The goal of this process is to compute a contrast and its variance from a GLM result before computing the student test. Drag and drop "Hb [Topo] | band(0.01-0.15Hz) | GLM OLS_precoloring fitted model" into the process field and press "Run". In the process menu, select "NIRS > GLM - 1st level contrast.
This process takes the contrast vector as input. You can either enter it as a SPM-like vector (eg '[ 1 -1]') or as a human-readable strings (eg '+ GraspingRight - Rest').
This will output a file called 'GLM con + GraspingRight - Rest' that contains the contrast matrix (eg cB) and its associated covariance matrix.
Drag and drop "GLM con + GraspingRight - Rest" into the process field and press "Run". In the process menu, select "NIRS > GLM - contrast t-test.
This will produce a file called 't-stat <> 0 : GLM con + GraspingRight - Rest'. To view the result, double click on this file. This will open a window showing the t-value contains into the file, you can then right-click on this figure and choose 'View topography'. You can then review the results for HbO or HbR by right-clicking on this new figure and selecting Montage > HbO > HbO or Montage > HbR > HbR as described in the next figure.
You can also use Brainstorm panel to choose the statistical threshold and the method for multiple comparison corrections that you want to use as described in the next figure.
Note: There are two major differences in the data processing if you are using pre-whitening :
- You should resample the data at 5hz. This is needed because the pre-whitening process can be very long to run when the sampling rate is high. (approximately 30 minutes for one subject of this dataset after the resampling)
- You should not use a low-pass filter as it will increase the auto-correlation of the noise. You can then replace the band-pass filter by a high-pass filter with a low cutoff frequency of 0.01Hz.
- From the menu of the Brainstorm main panel: File > Load protocol > Load from zip file > Select the downloaded protocol file: 'Nirstorm_workshop_PERFORM_2020/GLM_analysis/protocol_nirs_glm_group.zip'
To proceed to the group analysis, we will first do a group average by averaging each of the subject average. To do so, put every subject into the process field, and select the average file using the search option :
 you should end up with 6 files to average :
you should end up with 6 files to average :

The result is in the group analysis folder and can be analyzed as previously :

To perform group analysis using GLM, select all subject in the process field, and select 'GLM_OLS con' or 'GLM_white con' (if you want to use pre-coloring or pre-whitening) and compute NIRS > GLM - MFX group t-test.

We finally end up with this result at the group level using GLM and pre-coloring :

Note: In this process, we assume that the montage is consistent across the subject. (eg the montage is the same for each subject). This process will only keep the non-bad channel from all the subjects. If you are using a different montage for each subject or have missing channels, we recommend you to project the concentration variation on the cortex before doing the GLM.
This process is part of the experiment planning, taking place before the acquisition, where the experimenter wants to define where to place the optode on the scalp. Usually, a NIRS montage has a limited coverage over the head and hence optodes are placed specifically on top of a predefined target cortical region of interest (ROI). However, due to the non-trivial light propagation across head tissues as well as the complex cortical topology, the optode placement cannot be optimally based solely on a pure geometrical criterion.
Here, the computation of the optimal NIRS montage consists of finding the set of optode coordinates whose sensitivity to a predefined region of interest is maximized (Machado et al., 2014; Pellegrino et al., 2016).
To achieve this, this process requires:
- an anatomical MRI,
- pre-computed fluence data,
- a target ROI, either on the cortex or on the scalp.
For ease of configuration, the current tutorial relies on the Colin27 template adapted to NIRS (included in the workshop sample data). Indeed, fluence maps have already been pre-computed for this anatomy so that one can directly run the optimal montage process without going through the forward model computation. However, the most accurate results are obtained when using the subject-specific MRI. The current procedure can be straightforwardly transposed to any MRI.
Note that fluence data available in the downloaded sample data cover only the region of interest considered here. It amounts to around 1/10th of the head and data takes ~500Mb. In order not to have to download the whole head data (~6 Gb), NIRSTORM provides an on-demand fluence download system (specifically for the Colin27 template).
It will download only the fluence data required to cover the considered region if they are not already available on the local drive (in BRAINSTORM_USER_DIR/defaults/nirstorm/fluence/MRI__Colin27_4NIRS).
- Create a new subject, called "Subject_OM", with the default set-up
- Go to the anatomy view, right-click on Subject_OM and select "Use template > Colin27_4NIRS"
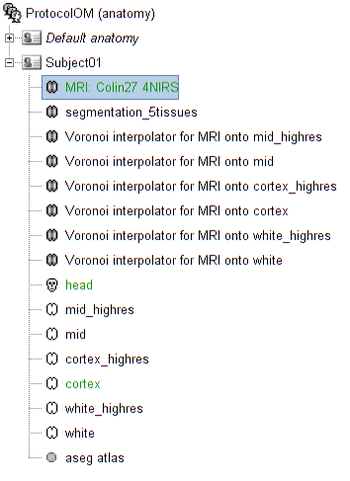
For ease of reproducibility, the ROI definition is atlas-based. To define the scout which represents this region of interest on which the sensitivity of the montage will be optimized, follow these steps:
- Open the mid cortical mesh by double-clicking on it in the anatomical view of Subject_OM
- Select the scout panel and use the drop-down menu to show the Destrieux atlas
- Select the scout defining the bottom of the frontopolar gyrus, labeled "RPF G_and_S_transv_frontopol".
- Click on "Scout > export to Matlab" and name it "scout_frontal"
- Use the drop-down menu to show "User scouts"
- Click on "Scout > Import" from Matlab and select "scout_frontal"
- Click on "Scout > Rename" and set the scout name to "scout_OM"
This ROI scout should end-up in the user-specific atlas "User scouts".

The optimal montage works by searching for the best possible layout of optode pairings withing a given region on the head mesh (search space). Each vertex in this region is a candidate holder on which an optode (source or detector) can be placed. A montage is the best if it maximizes the optical sensitivity over a pre-defined cortical region of interest.
Here, to avoid the explicit definition of the head region (search space), we will use the process "Compute optimal from cortex" which constructs the head scout by projecting the target cortical scout onto the head mesh. Alternatively, note that the head scout could be given explicitly while using the process "Compute optimal from head".
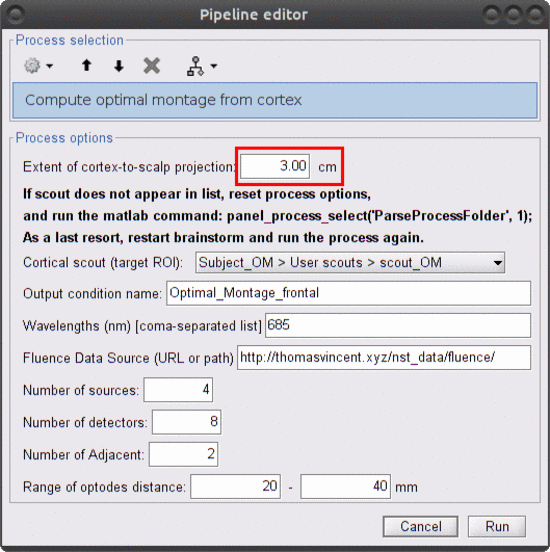
Without using any data, click on RUN (bottom left of the main brainstorm panel). Select the process "NIRS > Compute optimal montage from cortex". Here are the process parameters to fill in, along with some explanation about them:
-
Extent of cortex-to-scalp projection: 3 cm
This is the maximum distance used to project the cortical scout onto the head mesh to create the head scout (search space). More precisely, a head vertex will be part of the projected head scout if it is at most 3 cm away from the closest vertex in the cortical scout.
Make sure to use an extent of 3 cm since this value has to be consistent with the installed fluence files. -
Cortical scout (target ROI): Subject1 > User scouts > scout_OM
It defines the target ROI on which the sensitivity is optimized.
The selector is experimental. It may not reflect the actual list of scouts. There may also be an error regarding scout selection during the execution of the process. To solve this, quit the process, and try to: 1) Reset process options (see snapshot below) 2) Enterpanel_process_select('ParseProcessFolder',1)in the Matlab prompt, 3) restart brainstorm (if the two above did not work).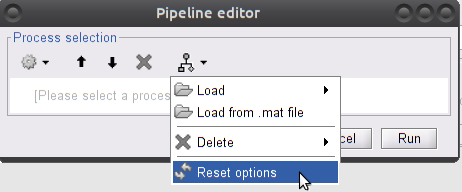
TODO PR for brainstorm menu item to reload processes (shortcut to
panel_process_select('ParseProcessFolder',1)Then restart the process again.
-
Output condition name: Optimal_Montage_frontal
The name of the condition (output functional folder) which will contain the computed montage. -
Wavelength (nm): 685
The wavelength of the fluence to load and which will be used to compute the optical sensitivity to the target cortical ROI. Note that optimal montages vary only very slightly across wavelengths. Hence we don't need to use the two actual wavelengths used during acquisitions. This saves some computation time! -
Fluence Data Source (URL or path): enter the path to the fluences included in the dataset you downloaded ( eg 'Nirstorm_workshop_PERFORM_2020/Module3_OptimalMontage/fluences_for_OM')
-
Number of sources: 4
Maximal number of sources in the optimal montage. The actual number in the resulting montage may be lower. -
Number of detectors: 8
Maximal number of detectors in the optimal montage. The actual number in the resulting montage may be lower. -
Number of adjacent: 2
Minimal number of detectors that must be seen by any given source. -
Range of optodes distance: 20 - 40 mm
Minimal and maximal distance between two optodes (source or detector). This applies to source-source, detector-dectector, and source-detector distances.
The optimal montage computations comprise the following steps with their typical execution time in brackets:
-
Mesh and anatomy loading [< 10 sec]
-
Loading of fluence files [< 1 min]
Fluence files will be first searched for in the folder .brainstorm/default/nirstorm/fluence. If not available, they will be requested from the resource specified previously (online here).
-
Computation of the weight table (summed sensitivities) [< 3 min]
For every candidate pair, the sensitivity is computed by summation of the normalized fluence products over the cortical target ROI. This is done only for pairs fulfilling the distance constraints.
-
Linear mixed integer optimization using CPLEX [~ 5 min]
This is the main optimization procedure. It explores the search space and iteratively optimizes the combination of optode pairings while guaranteeing model constraints.
-
Output saving [< 5 sec]
Switch to the functional tab of Subject_OM.
Under the newly created folder "Optimal_Montage_frontal", double click on "NIRS-BRS channels (19)". This will display the optimal montage along with some scalp regions. Ignore these regions for now.
In the view control panel, switch the "Surface" tab, click on the "+" icon in the top right and select "cortex". This will show the cortical mesh with the target frontopolar ROI for which the montage was optimized:

In order to use the optode coordinates in a neuronavigation, they can be exported as a CSV file.
Right-click on "NIRS-BRS channels (19)", select "File > Export to File" then for "Files of Type:" select "NIRS: Brainsight (*.txt)". Go to the folder Documents\Nirstorm_workshop_PERFORM_2020 and enter filename "optimal_montage_frontal.txt".
Here is the content of the exported coordinate file, whose format can directly be used with the Brainsight neuronavigation system:
# Version: 5
# Coordinate system: NIftI-Aligned
# Created by: Brainstorm (nirstorm plugin)
# units: millimetres, degrees, milliseconds, and microvolts
# Encoding: UTF-8
# Notes: Each column is delimited by a tab. Each value within a column is delimited by a semicolon.
# Sample Name Index Loc. X Loc. Y Loc. Z Offset
S1 1 61.801026 53.794879 20.827570 0.0
S2 2 58.614179 65.147586 -12.996690 0.0
S3 3 18.340509 84.492701 3.992621 0.0
S4 4 0.721182 85.338992 16.268421 0.0
D1 5 52.653816 67.712105 8.169515 0.0
D2 6 36.819009 72.131463 32.357219 0.0
D3 7 35.263908 77.335821 12.195818 0.0
D4 8 30.828533 79.924480 -26.315089 0.0
D5 9 43.354290 77.996386 -9.680977 0.0
D6 10 8.060217 87.471897 -13.272658 0.0
D7 11 -17.384161 84.440003 4.209488 0.0
D8 12 14.779627 78.451950 37.003541 0.0
Here is an example of optimal montage with a larger region: the right precentral gyrus.

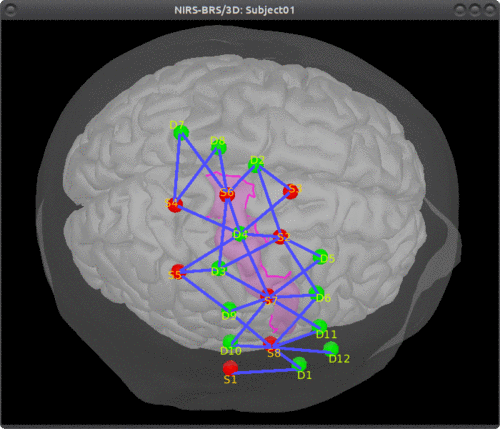
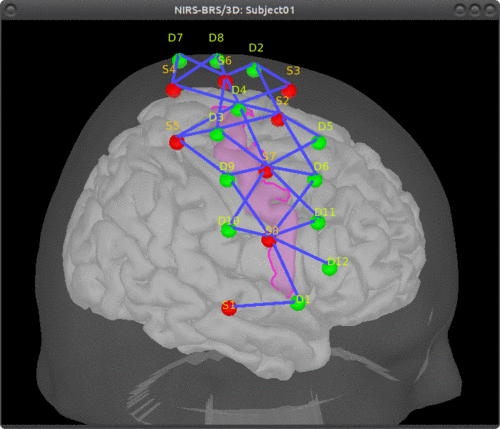
This tutorial illustrates how to perform NIRS reconstruction: estimate NIRS signals in the cortical layer that were at the origin of the signals measured in the channel-space on the scalp.
- One subject, one run of 19 minutes acquired with a sampling rate of 10Hz
- Finger tapping task: 20 blocks of 10 seconds each, with rest period of 30 to 60 seconds
- 3 sources, 15 detectors and 1 proximity detector
- 2 wavelengths: 685nm and 830nm
- 3T MRI anatomy processed by FreeSurfer 5.3.0 and SPM12
A complete data set with already pre-processed signals was downloaded in the sample data.
-
To load this data into brainstorm, from the menu of the Brainstorm main panel:
File > Load protocol > Import subject from zip and select the downloaded protocol file:
Nirstorm_workshop_PERFORM_2020/Module4_CorticalReconstruction/Subject_Reconstruction.zipThis will create the subject "Subject_Reconstruction" in the current protocol.
Switch to the "anatomy" view of the protocol. The anatomy data of subject Subject_Reconstruction mainly consists of:
- MRI: 3T MRI image of the subject
- Segmentation_5tissues: anatomical segmentation performed by FreeSurfer 5.3.0 and SPM12 tissues types contains scalp (labeled as 1), skull (labeled as 2), CSF (labeled as 3), gray matter (labeled as 4) and white matter (labeled as 5). In general, one could use segmentations calculated by other software. Note that the segmentation file was imported into Brainstorm as an MRI file, please make sure the label values are correct by double clicking on the segmentation file and reviewing using the tool MRIViewer of Brainstorm.
- Voronoi interpolator for MRI onto mid: interpolators to project volumic data onto the cortical mid surface. It will be used to compute NIRS sensitivity on the cortex from volumic fluence data in the MRI space. It has been computed using the nirstorm process "Compute Voronoi volume-to-cortex interpolator".
- scalp: scalp mesh calculated and remeshed by Brainstorm. To reduce the NIRS montage registration error, it is always better to remesh the head mesh so that two projected optodes do not end up on vertices that are too close.
-
mid: mid-surface calculated by FreeSurfer 5.3.0 which is in the middle of the pial and white surface. This is a resampled version of the original file provided by freesurfer, with a lower number of voxels to reduce the computation burden of the reconstruction. The NIRS signal spatial resolution is quite coarse anyway so we don't need many vertices. 25000 vertices is a fine recommendation for the reconstruction algorithm considered here.
Besides, note that the quality of this mesh is critical since it will the support of the reconstruction. Some mesh inconsistencies (crossing edges, non-manifolds, zero-length edges...) can occur during segmentation and have to been avoided or fixed.
The anatomy view should look like:
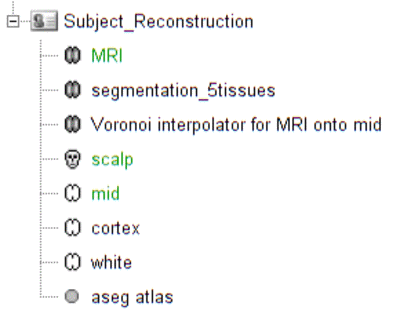
Verify that the default MRI, head mesh and cortical surface are the same as in this snapshot
The provided functional data is already preprocessed. Preprocessings were similar to the previous tutorial on channel-space analysis, except for the Modified Beer-Lambert Law (MBLL). The MBLL was here applied only partially without taking into account for the light path length correction. Indeed the cortical reconstruction process will take care of it, by explicitly modeling the light path through the head layers.

The functional view of the protocol consists of:
- FingerTapping_BlockDesign: contains the raw acquisition data.
-
FingerTapping_BlockDesign_dOD_SSR: contains delta OD data that has been preprocessed (e.g. filter, short distance channel regresion):
- NIRS-BRS sensors (90): NIRS channel file
- NIRS head model: pre-calculated NIRS reconstruction forward modle
- NIRS dOD: delta OD converted from raw data by doing logisim.
- NIRS dOD | low(0.1Hz): low-pass (0.1Hz high cut-off) filtered dOD to remove cardiac and respiratory artifacts
- FingerTapping (20 files): 20 block trials exported from NIRS dOD | low(0.1Hz) with a window size of -10 seconds to 35 seconds. These are peri-stimuli signal chunks.
- AvgStd FingerTapping (20): block-average of Finger tapping trials (20 files), with standard deviation. This pre-processed signal will be the input of the reconstruction.

The NIRS reconstruction process relies on the so-called forward problem which models light propagation within the head. It consists in generating the sensitivity matrix that maps the absorption changes along the cortical region to optical density changes measured by each channel (Boas et al., 2002). Akin to EEG, this is called here the head model and is already provided in the given data set (this is the item NIRS head model).
Note that Nirstorm offers head model computation by a calling a photon propagation simulation software: MCXlab. For additional details, please refer to this optional tutorial.
The visualize the current head model, first let's look at summed sensitivities which provide an overall cortical sensitivity of the NIRS montage. Double-click on Summed sensitivies WL1.
To go into more details, the head model can be explored channel-wise. Double click on Sensitivites - WL1 and Sensitivites - WL2. We used a trick to store pair information in the temporal axis. The two first digits of the time value correspond to the detector index and the remaining digits to the source index. So time=101 corresponds to S1D1. With the same logic, time=210 is used for S2D10. Note that some time indexes do not correspond to any source-detector pair and the sensitivity map is then empty at these positions. The following figure shows the summed sensitivity profile of the montage (bottom) and the sensitivity map of one channel S2D6 (top).

Reconstruction amounts to an inverse problem which consists in inverting the forward model to reconstruct the cortical changes of hemodynamic activity within the brain from scalp measurements (Arridge, 2011). The inverse problem is ill-posed and has an infinite number of solutions. To obtain a unique solution, regularized reconstruction algorithm are needed. In this tutorial, the Maximum Entropy on the Mean (MEM) will be used.
The MEM framework was first proposed by (Amblard et al., 2004) and then applied and evaluated in Electroencephalography (EEG) and Magnetoencephalography (MEG) studies to solve inverse problem for source localization (Grova et al., 2006; Chowdhury et al., 2013). Please refer to this tutorial for more details on MEM. This method has been adapted to NIRS reconstruction and implemented in nirstorm. Note that performing MEM requires the BEst – Brain Entropy in space and time package which is included in the Nirstorm package, as a fork.
To run the NIRS reconstruction using cMEM:
- Drag and drop AvgStd FingerTapping (20) into the process field
- Click "Run". In the process menu, select NIRS > Compute sources: BEst.

The parameters of this process are:
- Depth weighting factor for MNE: 0.3
-
Depth weighting factor for MEM: 0.3
These factors take into account that NIRS measurement have larger sensitivity in the superficial layer and are used to correct for signal sources that may be deeper. MNE stands for "Minimum Norm Estimation" and corresponds to a basic non-regularized reconstruction algorithm whose solution is used as a starting point for MEM. -
Set neighborhood order automatically: checked
This will automatically define the clustering parameter (neighborhood order) in MEM. Otherwise please make sure to set one in the MEM options panel during the next step.
Clicking on "Run" opens another parameter panel:

- Select cMEM in the MEM options panel. Keep most of the parameters by default and set the other parameters as:
- Data definition > Time window: 0-35s which means only the data within 0 to 35 seconds will be reconstructed
- Data definition > Baseline > within data checked, with Window: -10 to -0s which defines the baseline window that will be used to calculate the noise covariance matrix.
- Clustering > Neighborhood order: If Set neighborhood order automatically is not checked in the previous step, please give a number such as 8 for NIRS reconstruction. This neighborhood order defines the region growing based clustering in MEM framework. Please refer to this tutorial for details. If Set neighborhood order automatically is checked in the previous step, this number will not be considered during MEM computation.
Click "Ok". After less than 10 minutes, the reconstruction should be finished.
Unfold AvgStd FingerTapping (20) to show the MEM reconstruction results. To review the reconstructed maps, double click on the files cMEM sources - HbO and cMEM sources - HbR.
To show the reconstructed time courses of HbO/HbR, one need to define a region where to extract these time courses.
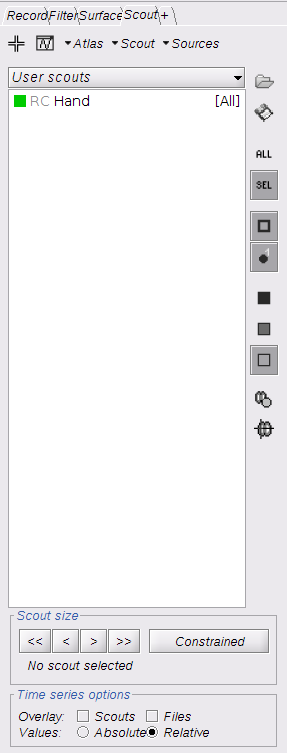
Switch to the "scout" tab in the view control panel. Then click on the scout "Hand". Under "time-series options" at the bottom, use "Values: Relative". Then in the "Scout" drop-down menu at the top of the tab, select Set function to "All". Finally, click on the wave icon to display the time courses.
By repeating the same process for both HbO and HbR, results should look like:

Nirstom also includes classical depth weighted Minimum Norm Estimate (MNE) to perform reconstitution. Please refer to this optional tutorial for details.
Fluences data reflecting light propagation from any head vertex can be estimated by Monte Carlo simulations using MCXlab developed by (Fang and Boas, 2009). Sensitivity values in each voxel of each channel are then computed by multiplication of fluence rate distributions using the adjoint method according to the Rytov approximation as described in (Boas et al., 2002). Nirstorm can call MCXlab and compute fluences corresponding to a set of vertices on the head mesh. To do so, one has to set the MCXlab folder path in matlab and has a dedicated graphics card since MCXlabl is running by parallel computation using GPU. See the MCXlab documentation for more details.
Drag and drop "AvgStd FingerTapping (20)" into the process field and press "Run". In the process menu, select "NIRS > Compute fluences for optodes by MCXlab". Enter the fluences output path as set the parameters as the following snapshot.

All these parameters are described in details on MCXLAB wiki page.
- The important ones are cfg.gpuid defining the GPU to use for calculation and cfg.nphoton which assigns the number of photons whose trajectories will simulated by Monte Carlo.
- If you have multiple GPUs, it is always better to use the one that is not working for displaying purpose.
- Swep GPU allows users to switch between GPUs to avoid too high temperature.
- To reduce the output file size, one could check the "Set threshold for fluences" and specify the threshold value.
Once you hit the "Run" button, Nirstorm will call MCXlab and calculate all the fluences and save them in the assigned folder. The next step is to calculate the head model - the sensitivity of each channel using fluences.
- Drag and drop "AvgStd FingerTapping (20)" into the process field and press "Run". In the process menu, select "NIRS > Compute head model from fluence".

- Copy and paste the fluence file path used previoulsy into the Fluence Data Source.
- Checkbox "Mask sensitivity projection in gray matter" allows to constrain sensitivity values to the gray matter to the cortex surface.
- "Smooth the surface based sensitivity map" will spatially smooth the sensitivity according to the FWHM value.
After running this process, a NIRS head model file called "NIRS head model from fluence" will be showed in the functional folder. Note that in Nirstorm, volumic sensitivity values were interpolated onto the cortical surface using the Voronoi based method proposed by (Grova et al., 2006). This volume to surface interpolation method has the ability to preserve sulco-gyral morphology.
To visualize the sensitivity or head model on the cortical space, one need to first extract the sensitivity data as functional data mapped onto the cortical mesh.
To do so, drag and drop "AvgStd FingerTapping (20)" into the process field and press "Run". In the process menu, select "NIRS > Extract sensitivity surfaces from head model". The sensitivity profile of each wavelength of the whole montage will be listed as "Summed sensitivites -WL1" and "Summed sensitivites -WL1". Users are also able to check the channel wised sensitivity map. Double click "Sensitivites - WL1" and "Sensitivites - WL2" and enter the number for instance "101" in the "Time panel" to show the sensitivity map of S1D1. With the same logic, 210 will be used for S2D10.
Minimum norm estimation (MNE) is one of the most widely used reconstruction method in NIRS tomography. It minimizes the l2-norm of the reconstruction error along with Tikhonov regularization (Boas et al., 2004).
Drag and drop "AvgStd FingerTapping (20)" into the process field and press "Run". In the process menu, select "NIRS > Source reconstruction - wMNE".
All the parameter definitions have the same meanning as in the MEM panel. Compute noise covariance of the baseline MNE will compute the noise covariance matrix according to the baseline window defined below. Otherwise, the identity matrix will be used to model the noise covariance.

Note that standard L-Curve method (Hansen, 2000) was applied to optimize the regularization parameter in MNE model. This actually needs to solve the MNE equation several times to find the optimized regularization parameter. Reconstruction results will be listed under "AvgStd FingerTapping (20)". Reconstructed time course can be plotted by using the scout function as mentioned in MEM section.

Boas D, Culver J, Stott J, Dunn A. Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head. Opt Express. 2002 Feb 11;10(3):159-70 pubmed
Boas D.A., Dale A.M., Franceschini M.A. Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy. NeuroImage. 2004. Academic Press. pp. S275-S288 pubmed
Hansen C. and O’Leary DP., The Use of the L-Curve in the Regularization of Discrete Ill-Posed Problems. SIAM J. Sci. Comput., 14(6), 1487–1503 SIAM
Arridge SR, Methods in diffuse optical imaging. Philos Trans A Math Phys Eng Sci. 2011 Nov 28;369(1955):4558-76 pubmed
Amblard C,, Lapalme E. and Jean-Marc Lina JM, Biomagnetic source detection by maximum entropy and graphical models. IEEE Trans Biomed Eng. 2004 Mar;51(3):427-42 pubmed
Grova, C. and Makni, S. and Flandin, G. and Ciuciu, P. and Gotman, J. and Poline, JB. Anatomically informed interpolation of fMRI data on the cortical surface Neuroimage. 2006 Jul 15;31(4):1475-86. Epub 2006 May 2. pubmed
Chowdhury RA, Lina JM, Kobayashi E, Grova C., MEG source localization of spatially extended generators of epileptic activity: comparing entropic and hierarchical bayesian approaches. PLoS One. 2013;8(2):e55969. doi: 10.1371/journal.pone.0055969. Epub 2013 Feb 13. pubmed
Machado A, Marcotte O, Lina JM, Kobayashi E, Grova C., Optimal optode montage on electroencephalography/functional near-infrared spectroscopy caps dedicated to study epileptic discharges. J Biomed Opt. 2014 Feb;19(2):026010 pubmed
Pellegrino G, Machado A, von Ellenrieder N, Watanabe S, Hall JA, Lina JM, Kobayashi E, Grova C., Hemodynamic Response to Interictal Epileptiform Discharges Addressed by Personalized EEG-fNIRS Recordings Front Neurosci. 2016 Mar 22;10:102. pubmed
Fang Q. and Boas DA., Monte Carlo simulation of photon migration in 3D turbid media accelerated by graphics processing units, Opt. Express 17, 20178-20190 (2009) pubmed
-
Tutorials:
-
Workshop and courses:
-
Manual:
-
Contribute:





