-
Notifications
You must be signed in to change notification settings - Fork 11
[WIP] GLM pipeline: surface and template based
The naming of subjects and condition folders is controlled by the function. To clearly separate the outputs of this function from other user-specific data, the suffix tag (__nspst_V1) is used. It is advised not to modify these folders. That way, one knows that the tagged data have been generated automatically by the pipeline function.
They can be safely removed and regenerated by running the script again.
The data set consists of one original subject undergoing a tapping task with an optimal montage over the motor cortex. This single subject has been duplicated 9 times to produce 10 dummy subjects coregistered with the Colin27 template. All dummy subjects have the same optode coordinates. Some noise was added in the 9 duplicated subjects to inject some variability.
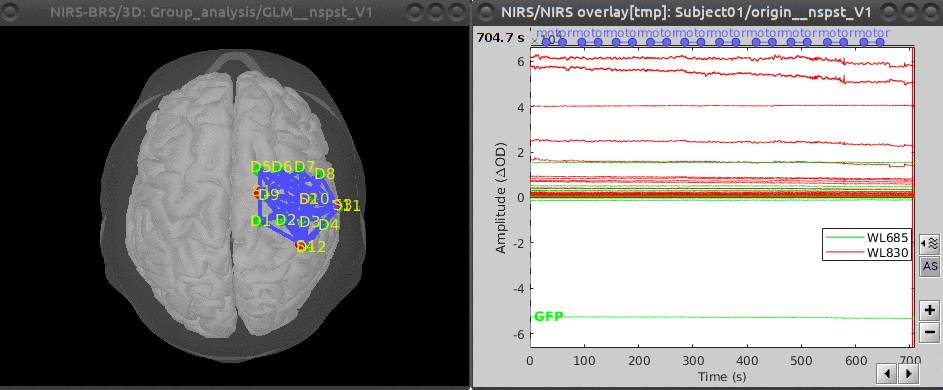
Here is the montage and signal plots for the initial subject:
This script provides a minimal working example of the pipeline on the set of 10 dummy subjects. The main steps are:
- importation of raw data files in brainstorm.
- default preprocessings up to cortical projection on the Colin27 template.
- a GLM with precoloring is run for all nodes of the surface to produce subject-level contrast maps.
- group-level GLM producing mixed effects contrast maps.
These steps are detailed in what follows.
The first part of the script takes care of insuring that brainstorm runs, setting the protocol and gathering sample data files. While customizing for your own study, adapt this part to build a list of nirs file names. Note that for every nirs file, a side-car file "optodes.txt" can be provided to specify the montage.
%% Setup brainstorm
if ~brainstorm('status')
% Start brainstorm without the GUI if not already running
brainstorm nogui
end
%% Check Protocol
protocol_name = 'TestSurfaceTemplateGroupPipelineV1';
if isempty(bst_get('Protocol', protocol_name))
gui_brainstorm('CreateProtocol', protocol_name, 1, 0); % UseDefaultAnat=1, UseDefaultChannel=0
end
% Set template for default anatomy
nst_bst_set_template_anatomy('Colin27_4NIRS_Jan19');
%% Fetch data
% Get list of local nirs files for the group data.
% The function nst_io_fetch_sample_data takes care of downloading data to
% .brainstorm/defaults/nirstorm/sample_data if necessary
[nirs_fns, subject_names] = nst_io_fetch_sample_data('template_group_tapping'); options = nst_ppl_surface_template_V1('get_options'); % get default pipeline options options is a struct gathering all default options, that can be customized afterwards.
sFiles = nst_ppl_surface_template_V1('import', options, nirs_fns, subject_names);This instruction imports all given nirs data files (nirs_fns) into brainstorm. The returned object sFiles is a cell array of strings gathering all imported brainstorm files. These files can then be customized before processings. See for instance the next step on event reading.
Note that an empty event group called "movement_artefacts" is created by the function. It can be filled by the user while manually tagging movement artefacts. It will be used afterwards during preprocessings to perform movement correction.
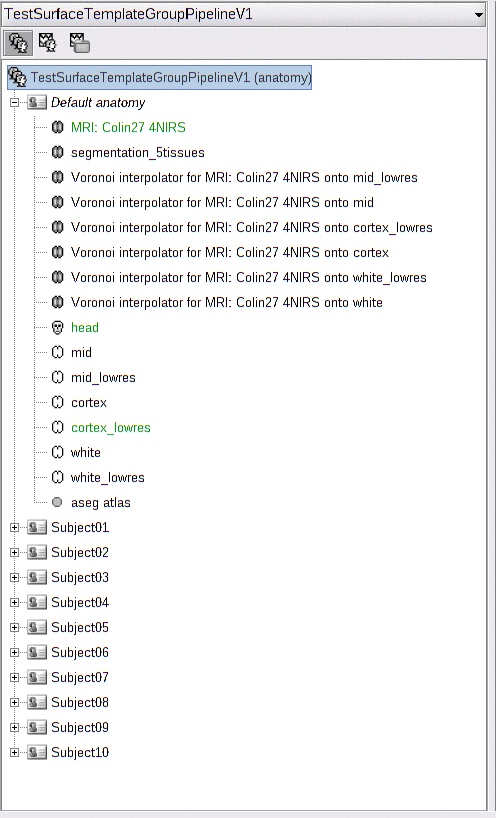
Here is the imported anatomy data. All subjects have the default anatomy: the Colin27 template.
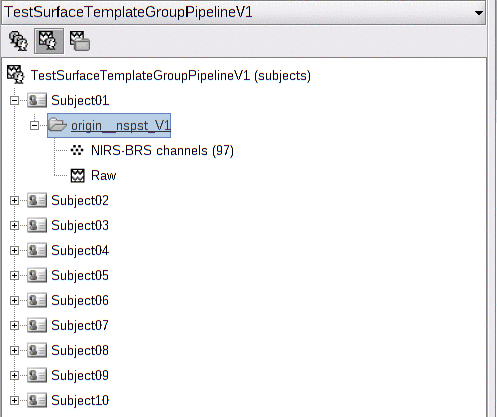
Here is the imported functional data. For every subject, the raw data is located in a folder called "origin".
In general, the stimulation events can be encoded in various ways, depending on the NIRS device or the experiment configuration. They can be bundled in the nirs files or stored in external files. So it is left to the user to load the events, just after importation.
In the current example, events are read from the 1st auxiliary channel, then renamed as "motor". The events duration is finally injected while converting to extended events. Here is the code:
% Read stimulation events from AUX channel
for ifile=1:length(sFiles)
evt_data = load(file_fullpath(sFiles{ifile}), 'Events');
if ~any(strcmp({evt_data.Events.label}, 'motor')) % Insure that events were not already loaded
bst_process('CallProcess', 'process_evt_read', sFiles{ifile}, [], ...
'stimchan', 'NIRS_AUX', ...
'trackmode', 3, ... % Value: detect the changes of channel value
'zero', 0);
% Rename event AUX1 -> motor
bst_process('CallProcess', 'process_evt_rename', sFiles{ifile}, [], ...
'src', 'AUX1', 'dest', 'motor');
% Convert to extended event-> add duration of 30 sec to all motor events
bst_process('CallProcess', 'process_evt_extended', sFiles{ifile}, [], ...
'eventname', 'motor', 'timewindow', [0, 30]);
end
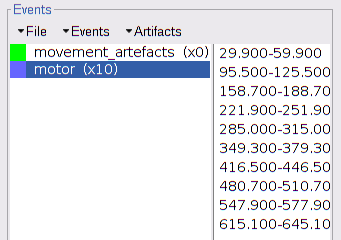
endHere are the loaded events: the motor ones, and the empty movement_artefacts group for later manual tagging:
The only mandatory parameters for the processing part of the pipeline are:
- the events to include in the design matrix of the GLM
- the contrasts.
Here the motor events are specified along with the motor contrast:
options.GLM_1st_level.stimulation_events = {'motor'};
options.GLM_1st_level.contrasts(1).label = 'motor';
options.GLM_1st_level.contrasts(1).vector = '[1 0]'; % a vector of weights, as a string Finally, all processings are run with:
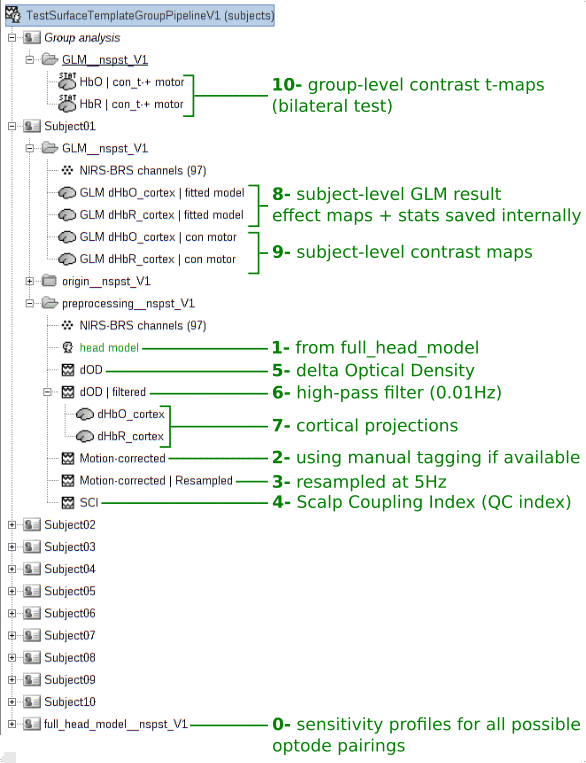
nst_ppl_surface_template_V1('analyse', options, subject_names); % Run the full pipelineHere is the commented illustration of the resulting brainstorm protocol after execution (the number at the beginning of each comment indicates the processing order):
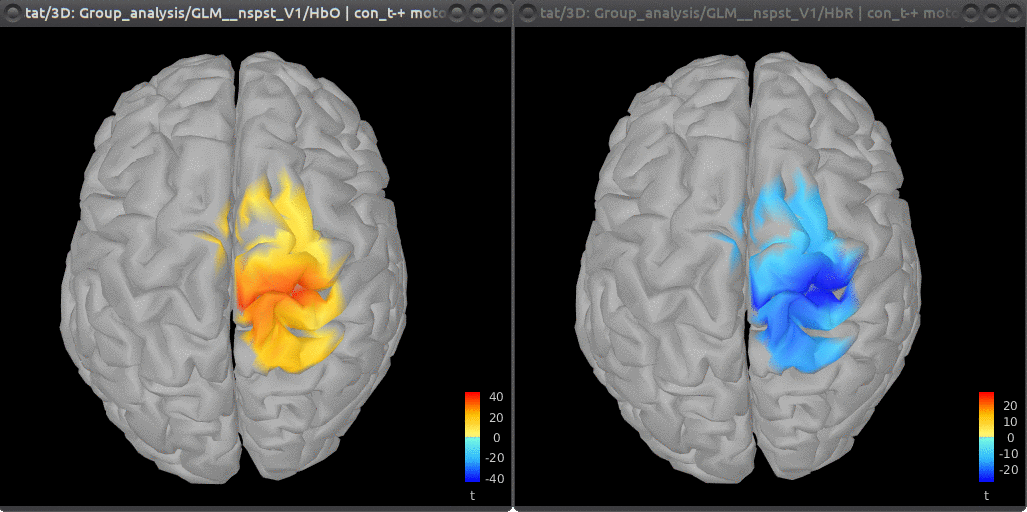
The group-level maps (HbO | con_t-+ motor and HbR | con_t-+ motor) thresholded at 0.05 with Bonferroni correction are:
-
Tutorials:
-
Workshop and courses:
-
Manual:
-
Contribute: