DIA-NN is an automated software suite for data-independent acquisition (DIA) proteomics data processing.
DIA-NN is built on the following principles:
- Reliability achieved via stringent statistical control
- Robustness achieved via flexible modelling of the data and automatic parameter selection
- Reproducibility promoted by thorough recording of all analysis steps
- Ease of use: high degree of automation, an analysis can be set up in several mouse clicks, no bioinformatics expertise required
- Powerful tuning options to enable unconventional experiments
- Scalability and speed: up to 1000 mass spec runs processed per hour
Download DIA-NN 2.0 for academic research: https://github.com/vdemichev/DiaNN/releases/tag/2.0.
DIA-NN 2.0 Enterprise for industry use: contact Aptila Biotech aptila.bio to purchase or obtain a trial license.
Installation
Getting started with DIA-NN
Changing default settings
Raw data formats
Spectral library formats
Sequence databases
Output
Command interface
Visualisation
Automated pipelines
PTMs and peptidoforms
Fine tuning prediction models
Multiplexing using plexDIA
Editing spectral libraries
Custom decoys
Integration with other tools
Quantification
Speed optimisation
Incremental processing
Basics of DIA data analysis
GUI settings reference
Command-line reference
Main output reference
Frequently asked questions
Support
Key publications
On Windows, download and run the .msi file. It is recommended to install DIA-NN into the default folder suggested by the installer. If your system may not have the common packages from Microsoft installed, download and run https://aka.ms/vs/17/release/vc_redist.x64.exe as well as https://go.microsoft.com/fwlink/?linkid=863265 and reboot your PC.
On Linux (command-line use only), download and unpack the Linux .zip file. The Linux version of DIA-NN is generated on Linux Mint 21.2, and the target system must have the standard libraries that are at least as recent. There is no such requirement, however, if you make a Docker or Apptainer/Singularity container image. To generate either container, we recommend starting with the latest debian docker image, the script make_docker.sh that does this is included. You can also check the excellent guide by Roger Olivella. For the best performance, use mimalloc with dynamic override as described here https://github.com/microsoft/mimalloc for all steps except predicted library generation. See Command interface for Linux-specific guidance.
It is also possible to run DIA-NN on Linux using Wine 6.8 or later.
If you are new to DIA proteomics, please take a look at the Basics of DIA data analysis.
DIA-NN analysis consists of two steps. First, use DIA-NN to generate a predicted spectral library from the sequence database, this library can then be reused for all experiments involving the respective organism. This first step is not necessary if you have a suitable empirical spectral library (e.g. generated by DIA-NN based on analysis of some DIA experiment). Second, have DIA-NN analyse the raw data with the selected spectral library. Each of the steps is set up in just several mouse clicks.
Predicted library generation:
- Click Add FASTA, add one or more sequence databases in UniProt format.
- Check FASTA digest checkbox. The Deep learning checkbox below gets checked automatically.
- (Optional) You can edit the Output library field. This is in fact a name 'template'. In workflows that generate empirical DIA-based libs, these are saved in the Apache .parquet format as specified by Output library. However, in this case DIA-NN will be generating a predicted library as specified by Predicted library below - it is the same name template, but the file has a different extension - .predicted.speclib, which is DIA-NN's own compact binary format for spectral libraries.
- Click Run. DIA-NN's log will be displayed in the log field (large field occupying the bottom-right part of the GUI window). Library generation takes about 3 minutes per million precursors on a 16-core desktop CPU.
Analysing with (any) spectral library:
- Click Spectral library and specify the library to use. Can be the .predicted.speclib as generated above, a .parquet lib generated by some previous DIA-NN analysis or some third-party predicted or empirical library in a compatible format (more about these below).
- Click Add FASTA, add one or more sequence databases in UniProt format, corresponding to the spectral library (i.e. if the library contains human proteins, any human database is fine here). If the library does not contain correct protein information (e.g. some third-party library), check Reannotate, this will update the protein information for each of the precursors in the library.
- Click Raw and select the raw data files. In case of Bruker timsTOF data, click instead .d (DIA) and specify the timsTOF acquisitions (which are folders with names ending with .d).
- (Optional) Adjust Main output. This is the name of the main output report generated by DIA-NN. It is also used by DIA-NN as the name template for any other report files it is going to generate (more about these below).
- Click Run. Note that DIA-NN will use the raw data to generate an empirical spectral library (file name specified by Output library).
If it is your very first time running DIA-NN, it makes sense to first explore how DIA-NN output looks like. For this, run DIA-NN on a dataset that can be analysed just in a couple of minutes. We recommend downloading raw data files (dia-PASEF 10ng.zip archive), empirical spectral library (K562-spin-column-lib.zip) and a FASTA database (human_canonical_uniprotkb_proteome_UP000005640_2023_12_16.fasta) from the Slice-PASEF benchmarks repository. When analysing this dataset, uncheck the MBR and Generate spectral library checkboxes.
Output:
-
The DIA-NN main report contains a list of all precursors ('Precursor.Id' column), with respective matched proteins ('Protein.Group' column) and quantities for both precursor and proteins ('Precursor.Normalised' and 'PG.MaxLFQ', respectively), for each run in the experiment. The report is in a .parquet format, it is basically a way to store text tables in a compact compressed format, it can be accessed with either R 'arrow' or Python 'pyarrow' or 'polars' packages.
-
In case you would like to immediately take a look at the protein quantities table, using software such as MS Excel, DIA-NN generates simple ready-to-use .pg_matrix.tsv and .unique_genes_matrix.tsv tab-separated tables of quantities.
The above information is sufficient for one to start using DIA-NN, it's indeed this easy! Please also check the Visualisation and Automated pipelines sections, these describe some very helpful functionality of the DIA-NN GUI, as well as Changing default settings below.
LC-MS-specific parameters. for publication-ready analyses, it is preferable to adjust the mass accuracies and the 'scan window' used by DIA-NN. These parameters provide guidance to DIA-NN, informing it on the expected magnitude of mass deviations (mass accuracies) and the expected number of DIA cycles per the average peptide elution time. By default, these are set to 0, meaning that DIA-NN will optimise them automatically for the first run in the experiment and then reuse the optimised settings for other runs. This optimisation process is 'noisy' in the sense that even replicate injections may not produce identical results. Therefore, the analysis results will depend on which run is first in the list of raw data files provided to DIA-NN. It's preferable to rather fix these three parameters to values know to be optimal for a particular LC-MS setup:
-
If the data were generated on timsTOF, set both Mass accuracy (MS/MS mass tolerance) and MS1 accuracy (MS1 mass tolerance) to 15.0 (ppm). Note that these settings provide a 'guidance' to DIA-NN, the actual m/z window used to match theoretical m/z values to the (m/z, intensity) pairs in the raw data file will be specific to both the precursor and the raw data in the vicinity of its putative retention time.
-
If the data were generated on Oritrap Astral, set Mass accuracy to 10.0 and MS1 accuracy to 4.0.
-
If the data were generated on TripleTOF 6600 or ZenoTOF, set Mass accuracy to 20.0 and MS1 accuracy to 12.0.
-
In other cases (e.g. Q Exactive and Exploris mass spectrometers) or if you would like to optimise everything to squeeze the best possible performance from the data, run DIA-NN on several representative runs (best to use any suitable empirical library, as this is the quickest) with Unrelated runs option checked and see what 'Optimised mass accuracy' and 'Recommended MS1 mass accuracy' it reports for those runs in the log.
-
Set Scan window to the approximate number of DIA cycles during the elution time of an average peptide. You can also have DIA-NN suggest the optimal one using an analysis with Unrelated runs checked.
How to interpret the rest of the present guide. The above covers DIA-NN use for 95% of projects. However, DIA-NN also offers powerful tuning options for fancy experiments. If you are just starting with DIA-NN, we recommend to first run it on a couple of simple datasets to get a feeling of how it works and then skim through the rest of this guide to see if any tips here can also be useful for your applications. The whole guide takes about 15 minutes to skim through and about 1-2 hours to study in detail.
In general, we recommended to:
-
Only change the default settings if (i) the change is recommended in the present guide, or (ii) you know that the change is necessary for your type of experiment and it's not covered in this guide, or (iii) you have already tested the default settings and you would like to see the effect of alternative settings.
-
Examine DIA-NN's log for warnings/errors. The warnings and errors are printed when they occur as well as at the end of the log, i.e. you can immediately see them once the DIA-NN analysis finishes.
-
DIA-NN prints some comments on the settings used at the top of the log, examine these the first time you use the settings to see if there are any recommendations.
The rest of this guide references extra options/commands you can pass to DIA-NN, please see Command interface for an introduction.
Formats supported: Sciex .wiff, Bruker .d, Thermo .raw, .mzML and .dia (format used by DIA-NN to store spectra). Conversion from any supported format to .dia is possible (except for Slice-type acqusitions on timsTOFs). When running on Linux (native builds, not Wine), only .d, .mzML, and .dia data are supported.
For .wiff support, download and install ProteoWizard - choose the version (64-bit) that supports "vendor files"). Then copy all files with 'Clearcore' or 'Sciex' in their name (these will be .dll files) from the ProteoWizard folder to the DIA-NN installation folder (the one which contains diann.exe, DIA-NN.exe and a bunch of other files).
Reading Thermo .raw files requires Thermo MS File Reader to be installed. It is essential to use specifically the version by the link above (3.0 SP3).
.mzML files should be centroided and contain data as spectra (e.g. SWATH/DIA) and not chromatograms.
Technology support
- DIA and SWATH are supported
- Acquisition schemes with overlapping windows are supported
- Gas-phase fractionation is supported
- Scanning SWATH and ZT Scan DIA are supported
- dia-PASEF/py-diAID are supported
- Slice-PASEF is supported
- midia-PASEF is supported
- Synchro-PASEF/VistaScan is supported, however accuracy of quantitation needs to be validated with benchmarks
- Orbitrap Astral is supported
- FAIMS with constant CV is supported
- multiplexing with non-isobaric tags and SILAC is supported
- MSX-DIA is not supported
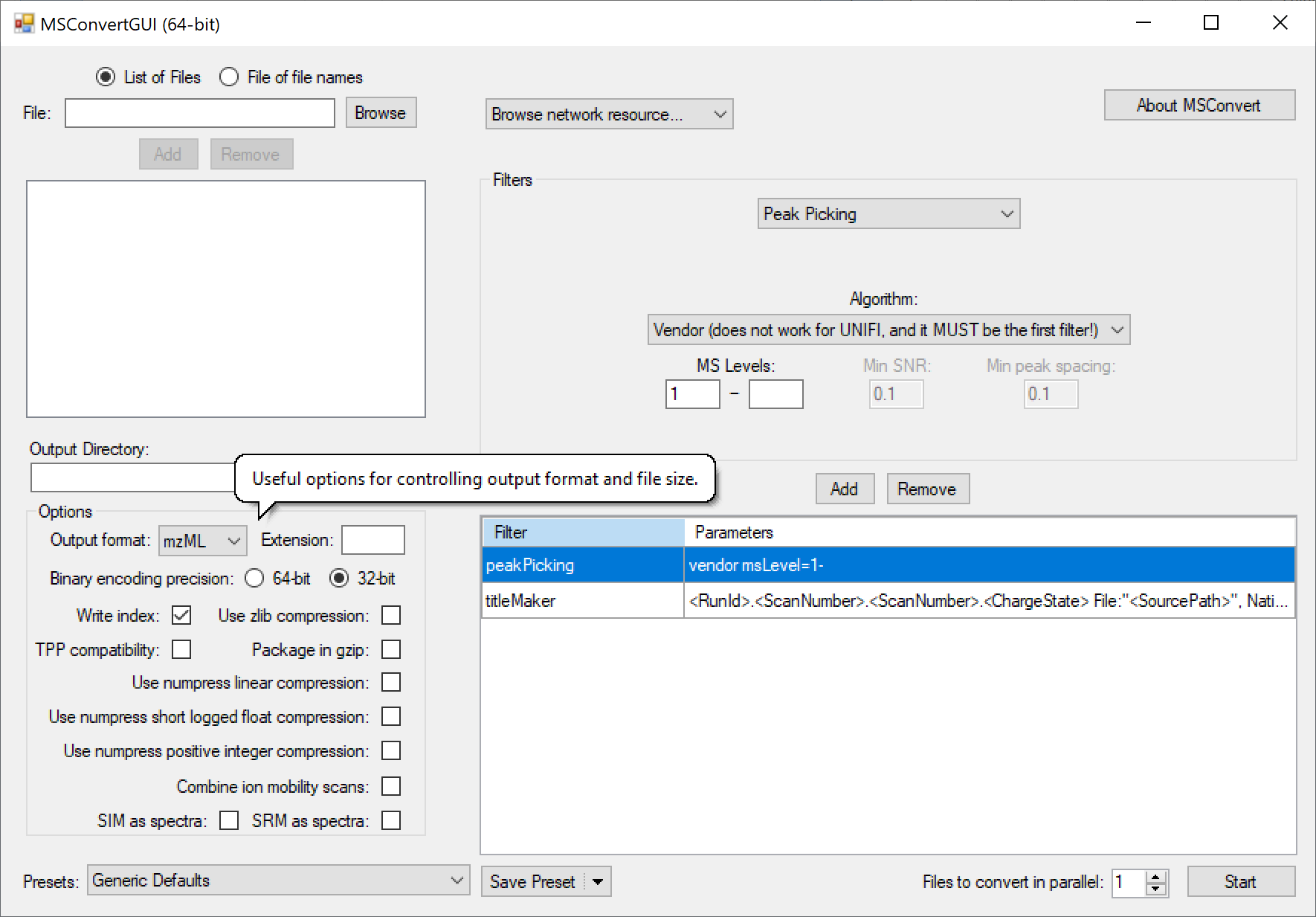
Conversion
Many mass spec formats, including those few that are not supported by DIA-NN directly, can be converted to .mzML using the MSConvertGUI application from ProteoWizard. This works for all supported formats except Bruker .d and SCIEX Scanning SWATH or ZT Scan DIA - these need to be accessed by DIA-NN directly. The following MSConvert settings must be used for conversion:
DIA-NN supports comma-separated (.csv), tab-separated (.tsv, .xls or .txt) or .parquet tables as spectral libraries, as well as .speclib (compact format used by DIA-NN).
In detail
Libraries in the PeakView format as well as libraries produced by FragPipe, TargetedFileConverter (part of OpenMS) are supported “as is”, however compatibility needs to be verified in each particular case (third party software version, its settings and the type of raw data).
DIA-NN can convert any library it supports into its own .parquet format. For this, click Spectral library (Input pane), select the library you want to convert, select the Output library file name (Output pane), click Run. If you use some exotic library format, it's a good idea to convert it to DIA-NN's .parquet and then examine the resulting library (using R 'arrow' or Python 'pyarrow' package) to see if the contents make sense.
All .tsv/.xls/.txt/.csv/.parquet libraries are just simple tables with human-readable data, and can be explored/edited, if necessary, using Excel or (ideally) R/Python. When editing and then saving DIA-NN's .parquet libraries, make sure to verify that the column types in the schema are unchanged: all floating point columns must be of the FLOAT parquet type and all integer columns must be of the INT64 parquet type.
DIA-NN accepts sequence databases in uncompressed FASTA format. The UniProt format is fully supported. For sequence databases in other formats, DIA-NN will most likely correctly extract the information on protein sequence IDs, however it may not correctly read the protein names, gene names and protein descriptions. In such cases, we recommend using any R package meant for handling FASTA databases to reformat the database in UniProt format.
The Output pane allows to specify where the output should be saved as well as the file names for the main output report and (optionally) the output spectral library. DIA-NN uses these file names to derive the names of all of its output files. Below one can find information on different types of DIA-NN output. For most workflows one only needs the main report (for analysis in R or Python - recommended) or the matrices (simplified output for MS Excel). When the generation of output matrices is enabled, DIA-NN also produces a .manifest.txt file with a brief description of the output files generated.
Main report
A text table in .parquet format containing precursor and protein IDs, as well as plenty of associated information. Please use either R "arrow" or Python "pyarrow"/"polars" packages to process. In addition to using R or Python, you can also view .parquet files with the TAD Viewer.
Most column names are self-explanatory, and the full reference as well as guidance on how to filter the main report can be found in Main output reference. The following keywords are used when naming columns:
- PG means protein group
- GG means gene group
- Quantity means non-normalised quantity
- Normalised means normalised quantity
- TopN means normalised protein quantity calculated using the TopN method when the Quantification strategy is set to Legacy (direct), using these is usually not recommended
- MaxLFQ means normalised protein quantity calculated using QuantUMS (Quantification strategy set to one of QuantUMS modes) or the MaxLFQ (Quantification strategy set to Legacy (direct)) algorithm, using quantities in this column is recommended for most applications
- Global refers to a global q-value, that is calculated for the entire experiment
- Lib refers to the respective value saved in the spectral library, e.g. Lib.Q.Value means q-value for the respective library precursor; when using MBR, Lib q-values will correspond to q-values in the empirical DIA-based lib created in the first pass of MBR
Matrices
These contain normalised QuantUMS or MaxLFQ quantities for protein groups ('pg_matrix'), gene groups ('gg_matrix'), unique genes ('unique_genes_matrix'; i.e. genes identified and quantified using only proteotypic, that is gene-specific, peptides) as well as normalised quantities for precursors ('pr_matrix'). They are filtered at 1% FDR, using global q-values for protein groups and both global and run-specific q-values for precursors. Additional 5% run-specific protein-level FDR filter is applied to the protein matrices, use --matrix-spec-q to adjust it. Sometimes DIA-NN will report a zero as the best estimate for a precursor or protein quantity. Such zero quantities are omitted from protein/gene matrices. Special phosphosite quantification matrices (phosphosites_90 and phosphosites_99 .tsv) are generated when phosphorylation (UniMod:21) is declared as a variable modification, see PTMs and peptidoforms.
Protein description
The .protein_description.tsv file is generated along with the Matrices and contains basic protein information known to DIA-NN (sequence IDs, names, gene names, description, sequence). Future versions of DIA-NN will include more information, e.g. protein molecular weight.
Stats report
Contains a number of QC metrics which can be used for data filtering, e.g. to exclude failed runs or as a readout for method optimisation. Note that the number of proteins reported here corresponds to the number of unique proteins (i.e. identified with proteotypic precursors) in a given run at 1% unique protein q-value. This number can be reproduced from the main report generated using precursor FDR threshold of 100% and filtered using Protein.Q.Value <= 0.01 & Proteotypic == 1. What's counted as 'protein' here depends on the 'Protein inference' setting.
PDF report
A PDF file with a number of QC plots for each run in the experiment, generated on Windows only. Extra QC plots will be added in future DIA-NN versions.
Flexible reanalysis
The Output pane allows to control how to handle the '.quant files'. Now, to explain what these are, let us consider how DIA-NN processes raw data. It first performs the computationally-demanding part of the processing separately for each individual run in the experiment, and saves the identifications and quantitative information to a separate .quant file. Once all the runs are processed, it collects the information from all the .quant files and performs some cross-run steps, such as global q-value calculation, protein inference, calculation of final quantities and normalisation. This allows DIA-NN to be used in a very flexible manner. For example, you can stop the processing at any moment, and then resume processing starting with the run you stopped at. Or you can remove some runs from the experiment, add some extra runs, and quickly re-run the analysis, without the need to redo the analysis of the runs already processed. All this is enabled by the Reuse .quant files option. The .quant files are saved to/read from the Temp/.dia dir (or the same location as the raw files, if there is no temp folder specified). When using this option, the user must ensure that the .quant files had been generated with the exact same settings as applied in the current analysis, with the exception of Precursor FDR (provided it is <= 5%), Threads, Log level, MBR, Cross-run normalisation, Quantification strategy and Library generation - these settings can be different. It is actually possible to even transfer .quant files to another computer and reuse them there - without transferring the original raw files. Important: it is strongly recommended to only reuse .quant files when both mass accuracies and the scan window are fixed to some values (non-zero), otherwise DIA-NN will perform optimisation of these yet again using the first run for which a .quant file has not been found.
Note: the main report in .parquet format provides the full output information for any kind of downstream processing. All other output types are there to simplify the analysis when using MS Excel or similar software. The numbers of precursors and proteins reported in different types of output files might appear different due to different filtering used to generate those, please see the descriptions above. All the 'matrices' can be reproduced from the main .parquet report, if generated with precursor FDR set to 5%, using R or Python.
DIA-NN is implemented as a graphical user interface (GUI), which invokes a command-line tool (diann.exe). The command-line tool can also be used separately, e.g. as part of custom automated processing pipelines. Further, even when using the GUI, one can pass options/commands to the command-line tool, in the Additional options text box. All these options start with a double dash -- followed by the option name and, if applicable, some parameters to be set. So if you see some option/command with -- in its name mentioned in this guide, it means this command is meant to be typed in the Additional options text box. Some of such useful options are mentioned in this guide, and the full reference is provided in Command-line reference.
When the GUI launches the command-line tool, it prints in the log window the exact set of commands it used. So in order to reproduce the behaviour observed when using the GUI (e.g. if you want to do the analysis on a Linux cluster), one can just pass exactly the same commands to the command-line tool directly.
diann.exe [commands]
Commands are processed in the order they are supplied, and with most commands this order can be arbitrary.
On Linux, the semicolon ';' character is treated as a command separator, therefore, ';' as part of DIA-NN commands (e.g. --channels) needs to be replaced with ';' on Linux for correct behaviour. Likewise, the exclamation mark '!' (e.g. in --cut) needs to be replaced with '!'.
For convenience, as well as for handling experiments consisting of thousands of files, some of the options/commands can be stored in a config file. For this, create a text file with any extension, say, diann_config.cfg, type in any commands supported by DIA-NN in there, and then reference this file with --cfg diann_config.cfg (in the Additional options text box or in the command used to invoke the diann.exe command-line tool).
DIA-NN provides two visualisation options.
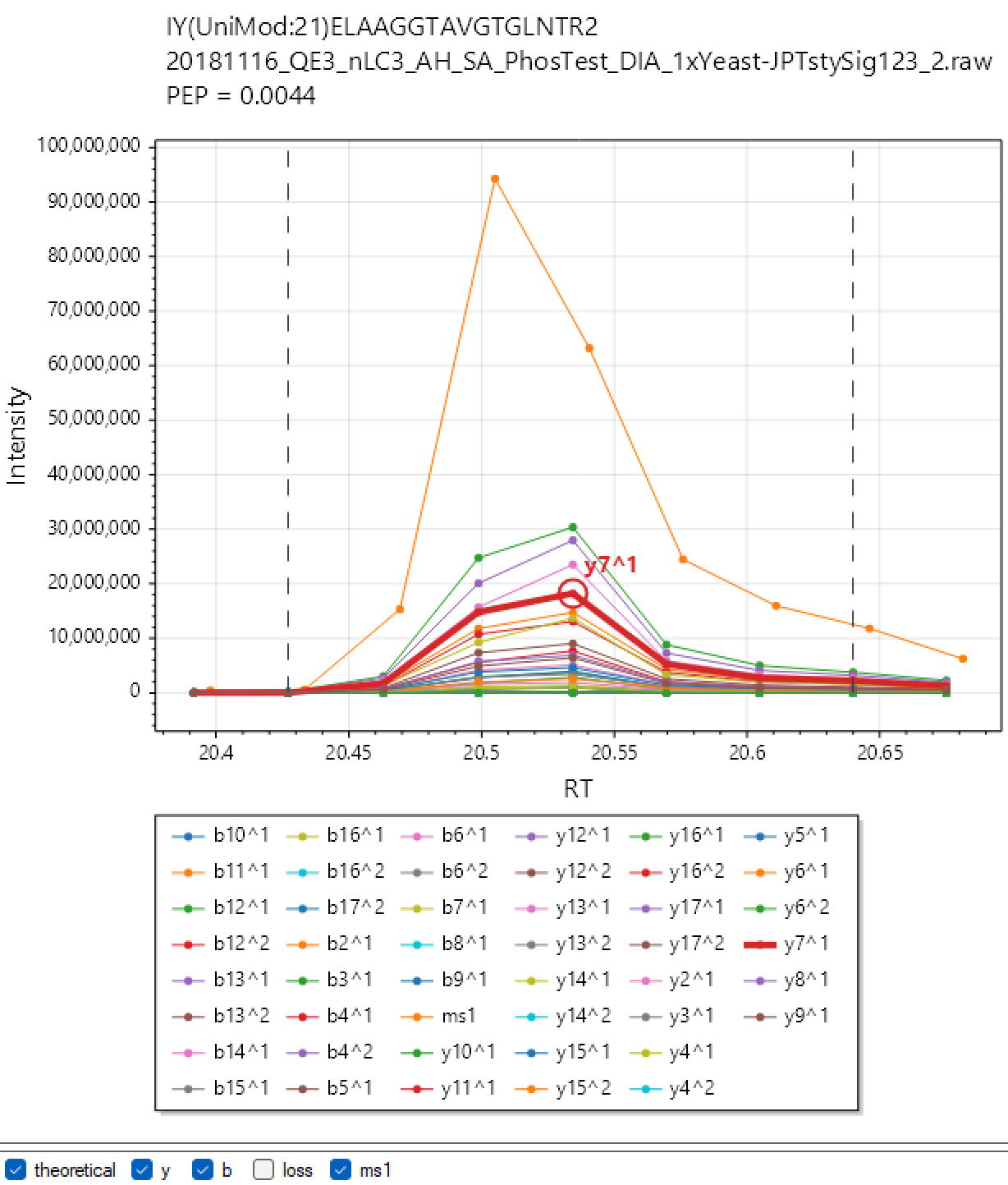
Skyline. To visualise chromatograms/spectra in Skyline, analyse your experiment with MBR and a FASTA database specified and then click the 'Skyline' button. DIA-NN will automatically launch Skyline. Currently this workflow does not support multiplexing and will not work with modifications in any format other than UniMod. Note: at the time of the release of DIA-NN 2.0 this functionality is not operational but is expected to work with Skyline versions to be released in the near future. This is related to the transition from .tsv to .parquet output in DIA-NN, which is not yet supported in Skyline.
DIA-NN Viewer. Analyse your experiment with the "XICs" checkbox checked and click the 'Viewer' button. By default "XICs" option will make DIA-NN extract chromatograms for the library fragment ions only and within 10s from the elution apex. Use --xic [N] to set the retention time window to N seconds (e.g. --xic 60 will extract chromatograms within a minute from the apex) and --xic-theoretical-fr to extract all charge 1 and 2 y/b-series fragments, including those with common neutral losses. Note that using --xic-theoretical-fr, especially in combination with large retention time window, might require a significant amount of disk space in the output folder. However the visualisation itself is effectively instantaneous, for any experiment size. Currently, DIA-NN Viewer does not support multiplexing, support will be added in future versions.
Note: The chromatograms extracted with "XICs" are saved in Apache .parquet format (file names end with '.xic.parquet') and can be readily accessed using R or Python. This can be sometimes convenient to prepare publication-ready figures (although can do that with Skyline or DIA-NN Viewer too), or even to set up automatic custom quality control for LC-MS performance.
Peptide & modification positions within a protein can be visualised using AlphaMap by the Mann lab https://github.com/MannLabs/alphamap.
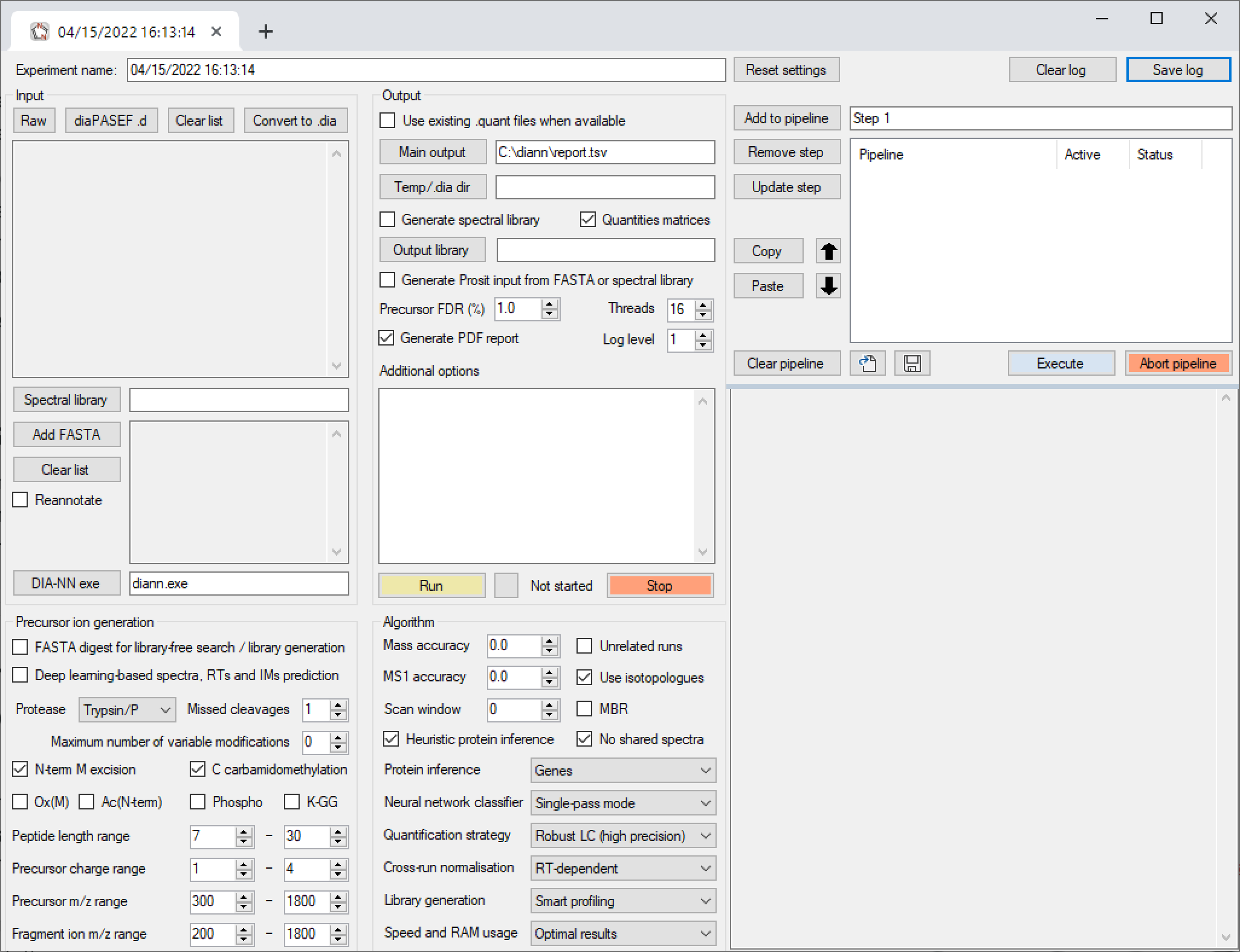
The pipeline window within the DIA-NN GUI allows to combine multiple analysis steps into pipelines. Each pipeline step is a set of settings as displayed by the GUI. One can add such steps to the pipeline, update existing steps, remove steps, move steps up/down in the pipeline, disable/enable (by double mouse-click) certain steps within the pipeline and save/load pipelines. Further, individual pipeline steps can be copy-pasted between different GUI tabs/windows (use Copy and Paste buttons for this). We always assemble all DIA-NN runs for a particular publication in a pipeline. One can also use DIA-NN pipelines to store configuration templates. When running the pipeline, each pipeline step is automatically saved as a separate one-step pipeline next to DIA-NN's main report.
The Apply RegEx functionality is useful primarily for migration of pipelines between different machines. This field allows to overwrite file paths in the pipeline using regular expression match-replace. The syntax is --regex "pattern to replace" "pattern to replace with",can use --regex multiple times. Use --ignore-case to ignore letter case when matching. For example, the command
--ignore-case --regex "diann.exe" "C:/DIA-NN/2.0/diann.exe" --regex "C:/Out" "C:/Out/2.0" --regex "C:\\Out" "C:/Out/2.0" --regex "mass-acc-cal 30" "mass-acc-cal 100"
changes the DIA-NN binary file location to that of a specific version, adjusts some of the file paths accordingly and adjusts the calibration mass accuracy setting if specified anywhere in the pipeline.
DIA-NN 2.0 introduces the ability to fine-tune its retention time (RT) and ion mobility (IM) prediction models. This is highly beneficial when looking for modifications on which DIA-NN's built-in models have not been trained on. Fine-tuning of the fragmentation predictor will also become available in the future.
Currently, the following modifications do no require any fine-tuning: UniMod:4 (C, carbamidomethylation), UniMod:35 (M, oxidation), UniMod:42 (N-term, acetylation), UniMod:21 (STY, phosphorylation). Fine-tuning may be somewhat beneficial for the following modifications: UniMod:121 (K, diglycine), UniMod:888 (N-term, K, mTRAQ), UniMod:255 (N-term, K, dimethyl). Fine-tuning is likely to signficantly boost detection of other modifications.
Tuning. To fine-tune DIA-NN's predictors, all you need is a spectral library (say, tune_lib.tsv, more on how to generate it yourself below) containing peptides bearing the modifications of interest. Type the following in Additional options and click Run:
--tune-lib tune_lib.tsv
--tune-rt
--tune-im
If the library does not contain ion mobility information, omit --tune-im. If some modifications are not recognised, declare them using --mod. Tuning usually takes several minutes. DIA-NN will produce three output files: tune_lib.dict.txt, tune_lib.tuned_rt.pt and tune_lib.tuned_im.pt. You can now generated predicted libraries using tuned models by supplying the following options to DIA-NN:
--tokens tune_lib.dict.txt
--rt-model tune_lib.tuned_rt.pt
--im-model tune_lib.tuned_im.pt
Generating the tuning library. If there is no suitable tuning library, one can always generate it directly from DIA data. For this, select one or several 'good' (typically, largest size) runs that are expected to contain peptides with the modifications of interest. These runs can also come from some public data set. Make a predicted library with DIA-NN by specifying all modifications of interest as variable or fixed (including those the predictor has already been trained on, if you expect to find them in the raw data). In vast majority of cases the max number of variable modifications can be set to 1-3, going higher is unlikely to be beneficial. Search the raw files using this predicted library in Proteoforms scoring mode, with Generate spectral library selected and MBR disabled. If the search space is large, the data comes from timsTOF, Orbitrap or Orbitrap Astral, and you would like to obtain the results quicker, set Speed and RAM usage to Ultra-fast. Optional: to optimise the performance, change the name of the output library and search the data again using --rt-window [X] and --im-window 0.2, where X is half of the gradient length (in minutes). Out of the two empirical libraries, choose the one with more modified peptides detected. Use this empirical library for fine-tuning.
If you use a tuning library that is not an empirical library generated by DIA-NN's lib-free search, and its RT & IM scales are significantly different from those produced by DIA-NN's predictors, it may be beneficial to adjust the RT & IM values in the tuning library to approximately match DIA-NN's (e.g. it's easy to do it if the tuning library contains some unmodified peptides, as those can be used for adjustment).
DIA-NN GUI features built-in workflows (Precursor ion generation pane) for detecting methionine oxidation, N-terminal protein acetylation, phosphorylation and ubiquitination (via the detection of remnant -GG adducts on lysines). Other modificaitons can be declared using --var-mod or --fixed-mod in Additional options.
Distinguishing between peptidoforms bearing different sets of modifications is a non-trivial problem in DIA: without special peptidoform scoring the effective peptidoform FDR can be in the range 5-10% for library-free analyses. DIA-NN implements a statistical target-decoy approach for peptidoform scoring, which is enabled by the Peptidoforms Scoring mode (Algorithm pane) and is also activated automatically whenever a variable modification is declared, via the GUI settings or the --var-mod command. The resulting peptidoform q-values reflect DIA-NN's confidence in the correctness of the set of modifications reported for the peptide as well as the correctness of the amino acid sequence identified. These q-values, however, do not guarantee the absence of low mass shifts due to some amino acid substitutions or modifications such as deamidation (note that DDA does not guarantee this either). They are also not a replacement for dedicated channel-confidence scores, see Multiplexing using plexDIA.
Note: for purely peptidomics applications, such as a typical phosphoproteomics experiment, we recommend the Proteoforms Scoring mode, see GUI settings reference for details.
Further, DIA-NN features an algorithm which reports PTM localisation confidence estimates (as posterior probabilities for correct localisation of all variable PTM sites on the peptide as well as scores for individual sites), included in the .parquet output report. The phosphosites_90 and phosphosites_99 .tsv files contain phosphosite-specific quantities, calculated using the Top 1 method (experimental), that is the highest intensity among precursors with the site localised with the specified confidence (0.9 or 0.99, respectively) is used as the phosphosite quantity in the given run. The 'top 1' algorithm is used here as it is likely the most robust against outliers and mislocalisation errors. However, whether or not this is indeed the best option needs to be investigated, which is currently challenging due to the lack of benchmarks with known ground truth. These matrices are currently not being produced when multiplexing is used (--channels), in this case please rely on the main report.
In general, when looking for PTMs, we recommend the following:
-
Essential: the variable modifications you are looking for must be specified as variable (via the GUI checkboxes or the Additional options) both when generating an in silico predicted library and also when analysing the raw data using any predicted or empirical library.
-
Settings for phosphorylation: max 3 variable modifications, max 1 missed cleavage, phosphorylation is the only variable modification specified, precursor charge range 2-3; to reduce RAM usage, make sure that the precursor mass range specified (when generating a predicted library) is not wider than the precursor mass range selected for MS/MS by the DIA method; to speed up processing when using a predicted library, first generate a DIA-based library from a subset of experiment runs (e.g. 10+ best runs) and then analyse the whole dataset using this DIA-based library with MBR disabled.
-
When the above succeeds, also try max 2 missed cleavages.
-
When looking for PTMs other than phosphorylation, in 95% of cases best to use max 1 to 3 variable modifications and max 1 missed cleavage.
-
When not looking for PTMs, i.e. when the goal is relative protein quantification, enabling variable modifications typically does not yield higher proteomic depth. While it usually does not hurt either, it will make the processing slower.
Of note, when the ultimate goal is the identification of proteins, it is largely irrelevant if a modified peptide is misidentified, by being matched to a spectrum originating from a different peptidoform. Therefore, if the purpose of the experiment is to identify/quantify specific PTMs, amino acid substitutions or distinguish proteins with high sequence identity, then the Peptidoforms (or Proteoforms) scoring option is recommended. In all other cases peptidoform scoring is typically OK to use but not necessary, and will usually lead to a somewhat slower processing and a slight decrease in identification numbers when using MBR.
Does DIA-NN need to recognise modifications in the spectral library?
Yes. If unknown modifications are detected in the library, DIA-NN will print a warning listing those, and it is strongly recommended to declare them using --mod. Note that DIA-NN already recognises many common modifications and can also load the whole UniMod database, see the --full-unimod option.
We have developed plexDIA based on DIA-NN, a technology that allows to benefit from non-isobaric multiplexing (mTRAQ, dimethyl, SILAC) in combination with DIA. To analyse a plexDIA experiment, one needs an in silico predicted or empirical spectral library. DIA-NN then needs to be supplied with the following sets of commands, depending on the analysis scenario.
Scenario 1. The library is a regular label-free library (empirical or predicted), and multiplexing is achieved purely with isotopic labelling, i.e. without chemical labelling with tags such as mTRAQ or dimethyl. DIA-NN then needs the following options to be added to Additional options:
- --fixed-mod, to declare the base name for the channel labels and the associated amino acids
- --lib-fixed-mod, to in silico apply the modification declared with --fixed-mod to the library
- --channels, to declare the mass shifts for all the channels considered
- --original-mods, to prevent DIA-NN from converting the declared modifications to UniMod
Example for L/H SILAC labels on K and R:
--fixed-mod SILAC,0.0,KR,label
--lib-fixed-mod SILAC
--channels SILAC,L,KR,0:0; SILAC,H,KR,8.014199:10.008269
--original-mods
Note that in the above SILAC is declared as label, i.e. it is not supposed to change the retention time of the peptide. It is also a zero-mass label here, as it only serves to designate the amino acids that will be labelled. What the combination of --fixed-mod and --lib-fixed-mod does here is simply put (SILAC) after each K or R in the precursor id sequence, in the internal library representation used by DIA-NN. --channels then splits each library entry into two, one with masses 0 (K) and 0 (R) added upon each occurrence of K(SILAC) or R(SILAC) in the sequence, respectively, and another one with 8.014199 (K) and 10.008269 (R).
Scenario 2. The library is a regular label-free library (empirical or predicted), and multiplexing is achieved via chemical labelling with mTRAQ.
Scenario 2: Step 1. Label the library in silico with mTRAQ and run the deep learning predictor to adjust spectra/RTs/IMs. For this, run DIA-NN with the input library in the Spectral library field, an Output library specified, Deep learning-based spectra, RTs and IMs prediction enabled, list of raw data files empty and the following options in Additional options:
--fixed-mod mTRAQ,140.0949630177,nK
--lib-fixed-mod mTRAQ
--channels mTRAQ,0,nK,0:0; mTRAQ,4,nK,4.0070994:4.0070994;mTRAQ,8,nK,8.0141988132:8.0141988132
--original-mods
Use the .predicted.speclib file with the name corresponding to the Output library as the spectral library for the next step.
Scenario 2: Step 2. Run DIA-NN with the following options:
--fixed-mod mTRAQ,140.0949630177,nK
--channels mTRAQ,0,nK,0:0; mTRAQ,4,nK,4.0070994:4.0070994;mTRAQ,8,nK,8.0141988132:8.0141988132
--original-mods
Note that --lib-fixed-mod is no longer necessary as the library generated in Step 1 already contains (mTRAQ) at the N-terminus and lysines of each peptide.
Scenario 3. The library is a regular label-free library (empirical or predicted), and multiplexing is achieved via chemical labelling with a label other than mTRAQ. The reason this scenario is treated differently from Scenario 2 is that DIA-NN's in silico predictor has not been specifically trained for labels other than mTRAQ, and therefore an extra step to generate predictions is not necessary. Simply run DIA-NN as you would do in Scenario 1, except the --fixed-mod declaration will have a non-zero mass in this case and will not be a label. For example, for 5-channel dimethyl as described by Thielert et al:
‐‐fixed‐mod Dimethyl, 28.0313, nK
--lib-fixed-mod Dimethyl
‐‐channels Dimethyl,0,nK,0:0; Dimethyl,2,nK,2.0126:2.0126; Dimethyl,4,nK,4.0251:4.0251; Dimethyl,6,nK,6.0377:6.0377; Dimethyl,8,nK,8.0444:8.0444
--original-mods
Scenario 4. The library is an empirical DIA library generated by DIA-NN from a multiplexed DIA dataset. For example, this could be a library generated by DIA-NN in the first pass of MBR (and you'd like to reuse it to analyse the same or some other runs). The Additional options will then be the same as in Scenario 1, Scenario 2: Step 2 or Scenario 3, except (important!) --lib-fixed-mod must not be supplied.
Scenario 5. The sample is a light sample with heavy spike-in proteins or peptides, that is only a small proportion of precursors should feature multiple channels. In this case, start with two unlabelled spectral libraries, one for the whole proteome, another for the spike-ins. Make sure that the RT and IM scales in those are similar and fragmentation information is likewise comparable - this will be the case if the libraries are predicted by DIA-NN. Using Editing spectral libraries make sure that (i) the whole proteome library does not contain precursors corresponding to spike-in peptides and (ii) the spike-in library has appropriate channel family tags applied to the precursor/peptide names. Combine the libraries in DIA-NN and proceed as in Scenario 3, except omit --lib-fixed-mod.
In all the scenarios above, an extra option specifying the normalisation strategy must be included in Additional options. This can be either --channel-run-norm (pulsed SILAC, protein turnover) or -channel-spec-norm (multiplexing of independent samples).
Output. When analysing multiplexed data, the main report in .parquet format needs to be used for all downstream analyses. Note that PG.Q.Value and GG.Q.Value in the main report are channel-specific, when using multiplexing. The quantities PG.MaxLFQ, Genes.MaxLFQ and Genes.MaxLFQ.Unique are only channel-specific if (i) QuantUMS is used and (ii) either the report corresponds to the second pass of MBR or MBR is not used.
Channel confidence. DIA-NN estimates channel-confidence of identifications (expressed as Channel.Q.Value and PG.Q.Value) by searching an in silico generated 'decoy' channel and then comparing the numbers of identifications in this channel and cognate channels, at a given score threshold. The decoy channel generated is displayed by DIA-NN at the top of its log and can be overriden using --decoy-channel. Note that the channel confidence estimates obtained will be biased if the decoy channel turns out to be a poor model for the target channel-mismatched identifications. For example, in case of +0, +4 and +8 target channels, decoy channel +12 will result in conservative channel q-values if +8 is a carrier while +0 and +4 are single cells, and in optimistic channel q-values if +0 is the carrier. However, this fluctations in accuracy of channel confidence estimation would typically still allow for quality quantification with any reasonable (0.01-0.05) q-value filters.
Note: QuantUMS quality metrics provide independent control for channel confidence and can be used in addition or instead of channel-specific q-value filtering.
This section summarises manipulations with spectral libraries that can be helpful for certain fancy experiments.
DIA-NN itself can:
- Convert any spectral library compatible with it to .parquet format. For this, specify the input library in Spectral library field and click Run (Raw field must be empty).
- Merge several .parquet or .tsv libraries, for this use multiple --lib commands in Additional options. Note the warnings printed by DIA-NN.
- Replace the spectra, RTs and IMs in the library with predicted ones using deep learning. Note --dl-no-fr, --dl-no-rt and --dl-no-im that allow to control what gets replaced.
- Apply fixed modifications to the library using --lib-fixed-mod. This changes peptide names and, if the modification has non-zero mass, also precursor and fragment masses.
Using R or Python, you can further:
- Edit .tsv or .parquet libraries in arbitrary way, including filtering, editing RTs and IMs, changing peptide sequences, adding 'faux' modifications (e.g. pasting '(SILAC)' after specific amino acids, to be later on used with --channels) and editing modification names, adding decoy peptides, marking fragments that should not be used for quantification, editing proteotypicity information for precursors, adjusting how proteins are annotated.
- Combine .tsv or .parquet libraries, here it is important to make sure that there are no duplicate precursors in the resulting library.
Note that if you edit and save a library in .parquet format to be used by DIA-NN, you need to ensure that (i) all floating point columns have the FLOAT parquet type and (ii) all integer columns have the INT64 parquet type.
This section describes DIA-NN's ability to customise its decoy models, this is not needed for 99.9% of experiments.
This functionality is howerver essential for correct handling of data wherein a substantial proportion of peptides incorporates specific sequence patterns, and, to our knowledge, is a unique feature of DIA-NN. We further plan to expand the ways in which decoy generation is controlled, up to supporting fully-custom decoy generation algorithms, based on the feedback from the proteomics community.
DIA-NN implements two main approaches to decoy generation based on a target peptide: (i) shuffling of the residues using a particular algorithm and (ii) mutation of a single residue. DIA-NN has the following decoy generation parameters, which it will attempt to abide by whenever possible and will only ignore if it cannot generate the decoy otherwise:
- --dg-keep-nterm [N] do not change the first N residues, default N = 1
- --dg-keep-cterm [N] do not change the last N residues, default N = 1
- --dg-min-shuffle [X] aim for any fragment mass shift introduced by shuffling to exceed X in absolute value, default 5.0
- --dg-min-mut [X] aim for the precursor mass shift during mutation to be at least X in absolute value, default 15.0
- --dg-max-mut [X] aim for the precursor mass shift during mutation not to exceed X in absolute value, default 50.0
In particular, the options restricting N-term and C-term residue changes are essential in a situation when the target peptides are e.g. synthetic peptides that all share several N-term or C-term residues.
This is a quick reference section for third-party software developers.
Starting with DIA-NN 2.0, it is possible to supply DIA-NN with decoy peptide queries in the spectral library (.parquet) along with the target peptides. This allows DIA-NN to use its MBR-optimised algorithm to correctly control FDR with empirical libraries generated by third-party software. For this, the third-party software needs to generate its libraries in DIA-NN-compatible .parquet format (i.e. include all the columns, the column 'Flags' can have 0 values). Ideally, the libraries should contain:
-
A proportion of decoy peptides, corresponding the to q-value filtering applied when generating the library. DIA-NN will search these decoys in addition to the regular decoys it generates. Therefore, if the fraction of decoys is in the range of tens of percent (i.e. library FDR is >= 0.1), this will make the resulting DIA-NN's FDR estimates too conservative, which is fine for most experiments. Nevertheless, this ensures correct FDR control even with libraries filtered at >= 0.5 q-value.
-
Q-values for all entries, target and decoy. While this is not essential to ensure FDR control provided decoys are included, DIA-NN's algorithms use these q-values to improve identification performance. In case decoys are not provided, including q-values may, in most cases, largely ensure correct FDR control by itself. It is, therefore, always recommended.
The numeric columns in DIA-NN's .parquet libraries are of types INT64 and FLOAT, other types should not be used.
For third-party downstream tools, it may be useful to have DIA-NN also export the decoy identifications using --report-decoys.
DIA-NN implements Legacy (direct) and QuantUMS quantification modes. The default QuantUMS (high-precision) is recommended in most cases. QuantUMS enables machine learning-optimised relative quantification of precursors and proteins, maximising precision while in many cases eliminating any ratio compression, see Key publications. DIA-NN 2.0 has a much improved set of QuantUMS algorithms, compared to our original preprint.
Note that if you are analysing with an empirical library, you can quickly generate reports corresponding to different quantification modes with Reuse .quant files. This can also be done just for a subset of raw files, i.e. if you have analysed also blanks and now wish to exclude them.
We have observed that:
-
QuantUMS performance is largely unchanged regardless of the experiment size, i.e. it is suitable for large experiments.
-
QuantUMS works well also on experiments which include very different sample amounts (tested with 10x range across different samples). Note, however, that in this case the optimisation of QuantUMS parameters, automatically performed by DIA-NN, largely tunes the algorithm to quantify most accurately 'representative' identifications, i.e. if the experiment consists of 10 bulk runs and 10 single cell runs, optimisation will turn out to be optimal for bulk runs.
-
QuantUMS has been tested with sub 2-minute to over 90 minute gradients and with both bulk and single cell-like samples.
-
QuantUMS is particularly beneficial for Orbitrap Astral data due to the high quality of its MS1 spectra.
Below are some recommendations for the use of QuantUMS.
High-accuracy. Use the high-accuracy mode when you'd like to minimise the ratio compression at the cost of precision. Note that while it does a very good job at this in most cases, DIA-NN still cannot completely eliminate ratio compression for low-abundant precursors and proteins in challenging samples, e.g. when analysing nanogram amounts at 200-500 samples/day throughput. It is easy to see why: many precursors in such cases will not have a single high quality signal recorded.
For large experiments, train QuantUMS on a subset of runs (e.g. 10 to 100 representative runs with medium to high numbers of identified precursors) and then quantify the whole experiment by reusing the QuantUMS parameters. For this, first run QuantUMS on a subset of runs, you can do this by just selecting only those runs in Raw files window and checking Reuse .quant files. Alternatively, you can specifically instruct QuantUMS to train only on a range of raw files (by index, starting with 0) using --quant-train-runs first:last, e.g. --quant-train-runs 0:5 will perform training on identifications from the first 6 runs. A third option (not recommended) is to instruct DIA-NN to automatically select N runs (use N between 6 and 100) to train QuantUMS with --quant-sel-runs [N]. As output, you will see a list of quantificaton parameters printed in the log, e.g.
Quantification parameters: 0.330076, 0.123932, 1.03485, 0.324712, 0.241882, 0.271245, 0.161143, 0.0203897, 0.019745, 0.0728814, 0.0574469, 0.0620889, 0.206804, 0.0761579, 0.0977414, 0.0613432
You can then use those parameters and skip training on the whole (large) experiment by using --quant-params, e.g.
--quant-params 0.330076, 0.123932, 1.03485, 0.324712, 0.241882, 0.271245, 0.161143, 0.0203897, 0.019745, 0.0728814, 0.0574469, 0.0620889, 0.206804, 0.0761579, 0.0977414, 0.0613432
QuantUMS can be trained on replicate injections of the same sample: we have verified on a diverse set of LC-MS setups that such training results in near-optimal parameters even when trained on triplicates. Nevertheless, QuantUMS does require at least some degree of variation in the data. Even in replicate injections, some quantitative variation is always present due to flucations in instrument performance. Still, we recommend training QuantUMS on subsets of runs that include at least some non-replicates. This is no limitation, given that any 'real' experiment encompasses biological variation.
How optimal specific parameters are for a particular experiment depends on the LC-MS settings (gradient, acc/ramp times, acquisition scheme) and the general type of sample (e.g. whole-cell vs AP-MS). It is likely OK to even reuse QuantUMS parameters between experiments with the same LC-MS settings and general sample types, so long as the training is performed on data corresponding to the same or larger sample amounts and the 'instrument sensitivity' was at its highest when the training data were acquired. The opposite is not recommended.
Filtering. One of the benefits of QuantUMS is that it provides 'quality' metrics for each quantity it reports. We recommend using both Quantity.Quality and PG.MaxLFQ.Quality for filtering. In most cases, it makes sense to apply a relatively strict filter on the average quality metric for the analyte and much less strict filter on its run-specific quality metric. The average filter may increase numbers of DE analytes at a given FDR, as less analytes under consideration mean less strict multiple testing correction. The run-specific filter is best chosen to retain the vast majority of identifications, while eliminating the ones with very low quality quantities.
Legacy mode is to be used in the following cases:
-
Always use the legacy mode when benchmarking LC-MS (settings). This is due to the fact that QuantUMS does not have the objective to minimise CV values, rather its goal is to achieve a good balance between precision and accuracy. This balance may differ significantly between different LC-MS settings, and using a single metric such as the median coefficient of variation (CV) reflects only part of this balance. Therefore, for experiments where QuantUMS simply decides to emphasise precision to a greater extent, CV values will appear better.
-
You would like to use the top N protein quantities (normally not recommended) instead of the built-in QuantUMS MaxLFQ-like algorithm, QuantUMS precursor quantities are currently not inteded to be used for top N as QuantUMS is inherently a 'relative quantification' method, whereas top N effectively implies comparing quantities of different precursor ion species.
-
For the same reason, use the legacy mode if you would like use the quantities stored in the phosphosite matrices produced by DIA-NN, as these are generated using the top 1 method. These matrices are still being saved in the QuantUMS mode, however in this case they are only good as a quick reference of whether a site was identified. You can still perform phosphosite quantification using a MaxLFQ algorithm (in R) based on DIA-NN's QuantUMS precursor quantities in the main report.
-
For the same reason, use the legacy mode for IBAQ.
-
In all the above (rare) cases where comparisons between quantities of different precursor species are performed (= absolute quantification is important) and the legacy mode is therefore appropriate, also consider just using the raw fragment quantities (at the apex), reported by DIA-NN when using --export-quant, e.g. summing the first 3 fragments may in some cases be better than using the legacy quantities. The normalisation factors calculated by DIA-NN in any mode can also be applied to the raw fragment quantities. Further, in all such scenarious you may want to use the top N approach (with N between 1 and 3) for any kind of precursor quantities aggregation (e.g. protein quant) rather than MaxLFQ (because if MaxLFQ were appropriate, would be better to use QuantUMS).
Zero quantities. Any of the quantities produced by DIA-NN may be equal to zero. The best interpretation here is that such quantities likely indicate an analyte with low concentration. If you would like to log-transform the data, can just replace those with NAs. We prefer to report zeroes rather than change the algorithm to output noise or NAs, as zeroes here provide information which may be of value for some downstream workflows.
Synchro-PASEF. When analysing Synchro-PASEF data, use --quant-tims-sum, which makes DIA-NN quantify each fragment in each DIA cycle by summing the signals recorded in all cognate frames within the cycle. While this is likely to provide fairly good performance, it is unknown if minor changes in the IM calibration of the instrument may add to batch effects when analysing using Synchro-PASEF, we will have certainty once benchmarks are available to assess this.
Normalisation. DIA-NN implements different normalisation modes. Normalisation can correct for different amounts of input material and sample losses. The RT-dependent normalisation further corrects for factors that differentially affect peptides depending on their hydrophobicity (reflected by their RT), i.e. some types of sample preparation losses, including desalting, as well as possibly also fluctations in the ion source performance. There are multiple examples of experiments where RT-dependent normalisation has a major positive effect on quantiative performance and no known cases where it is substantially detrimental.
Limitations of normalisation. In general, any kind of generic normalisation in proteomics only makes sense under the following assumption: when only considering 'biological' variability of interest, most of the peptides are not differentialy abundant, or the numbers of upregulated / downregulated (in comparison to experiment-wide averages) peptides in each sample are about the same. This assumption often is not strictly satisfied. Reasons range from non-linear dynamic range of the LC-MS to fundamental biological differences between the samples (e.g., ideally, different normalisation factors may need to be applied to cytosilic, plasma membrane or chromatin proteins, when comparing two single cells of different sizes). However, in many 'typical' proteomics experiments the normalisation algorithms implemented by DIA-NN work very well still. Nevertheless, it is recommended to not include blanks/failed runs in you quantitative analyses, i.e. if you are only processing blanks to figure out how many proteins are detectable in these, you can always process them separately with the emprical library produced by DIA-NN based on your experiment. Further, best to avoid a situation when the majority of precursors in a particular sample are not detectable in most other samples, e.g. not to include bulk runs in the quantitative analyses of single cell samples (but can absolutely use them to generate the empirical library).
Disabling normalisation. Given the above, there are scenarious (e.g. AP-MS, any kind of protein fractionation, time-series tracking isotopic label incorporation) when it may be desirable to apply custom normalisation that takes into account the nature of the experiment. In this case, we suggest either (i) disabling the normalisation in DIA-NN - you can still use MaxLFQ quantities in this case or (ii) keeping normalisation set to RT-dependent and applying custom normalisation on top of that, to benefit from DIA-NN correcting for any RT-dependent perturbations. The latter makes sense if there is still a considerable fraction of peptides that are shared between the samples.
Fold-changes. Given that normalisation is never 100% perfect, it is usually recommended to incorporate fold-change cutoffs in any differential expression (DE) analysis. These do not need to be particular large in magnitude, but should at least exceed the expected error in normalisation, e.g. when treating a cell line with drug compounds that have minor effects on the cells, even 10% fold-change cutoff may be appropriate. Fold-change cutoffs have also a further benefit of substantially reducing the FDR. Important: fold-change cutoffs must be applied after adjusting the p-values via multiple testing correction, not before.
Detecting normalisation errors. Errors in normalisation may be easy to identify. For example, the number of points with positive and negative fold-changes on a volcano plot should almost always be balanced. Indeed, one or several pathways may be upregulated in one condition compared to another, but if most pathways seem to be upregulated, it would mean most proteins are upregulated, implying that the dataset has not been correctly normalised. This does not apply to some special cases like AP-MS though.
A simple normalisation test can be carried in R or Python. For any two samples
- Calculate log2 fold-change of each analyte (precursor or protein) between the samples.
- Plot the histogram of the above log2 fold-changes.
- Indicate the median and half sample mode (use hsm() function of the modeest R package) on the histogram. Both these values should be close to 0.
This section focuses on factors that allow to increase the speed of processing and reduce RAM usage with large predicted spectral libraries.
Analysing a subset of runs. DIA-NN is very good at creating quality emprical spectral libraries from DIA data, and it does not need the whole experiment to do this. Although DIA-NN has been successfully used to search tens of thousands of runs lib-free, this is not really necessary in 99.9% of cases. In contrast, when processing large experiments, we recommend selecting 20 to 100 high quality runs (often selecting just the largest files works well) and create an empirical library from those (do not include blanks or failed runs here, they take by far the longest to process and are useless for library creation). This library can then be used to search the entire experiment (with MBR off). Further, if you have just acquired an experiment and want to quickly confirm that e.g. the runs did not fail and the mass calibration is OK, any suitable library can be used for this purpose, e.g. any public library or a DIA-based empirical library created based on a single run.
Reducing the search space. The time to search a file with a large library is approximately proportional to the size of the library. Therefore, we recommend to strictly follow the recommendations in this guide with respect to specifying variable modifications (i.e. only specify them if there are compelling reasons for this), at least for the first analysis. Once you have the data obtained using recommended settings, can see if including extra modifications improves identification numbers (in vast majority of cases it does not).
Reducing RAM usage. DIA-NN requires just under 0.5Gb RAM to store 1 million library precursors. That is a 3-million human tryptic digest library will require 1.5Gb RAM, while a 50-million library for phosphoproteomics will require about 25Gb of RAM. RAM is further used to store the raw data file that is being processed and for temporary storage of candidate PSMs. The requirements of the latter can be minimsed by adjusting Speed and RAM usage. There is currently a limit of max 1 billion precursor ions in the library, while DIA-NN is fine searching anything below that, provided it has enough RAM. If you are dealing with huge sequence databases, ubiquitous variable modifications or non-specific digests, resulting in libraries containing hundreds of millions precursors, it may make sense to split them and search separately. For example, a charge 2 length 8-13 (primary range for HLA peptides) non-specific human digest library (see --cut) is roughly 97 million precursors, i.e. including other charge states and the full length range here would not be manageable on regular desktop PCs. However, it is OK to search charge 1 and charge 2 separately, and then combine the resulting empirical libraries for the quantitative analysis. Similarly, predicted libraries can be also split based on the precursor m/z ranges. Further, imagine a metaproteomics experiment that is to be searched against 1000s of species. In this case, it may make sense to split the species into groups (e.g. by taxa or even randomly) and search those groups separately, looking, say, only for charge 2 precursors in 8-20 length range. This will allow to identify any confidently detected species (e.g. at least N proteins detected using proteotypic peptides) and then research only those in one go.
Speed and RAM usage. This setting is useful on some instruments. Specifically, we have observed on a number of experiments that with MBR the peptidoform-confident identification numbers obtained in Ultra-fast mode can be the same or almost the same as in the Optimal results mode, while the Ultra-fast mode is often fold-change faster. This in particular applies to fairly heterogeneous samples (most real experiments). Therefore, on samples that take very long time to process (e.g. long-gradient slice/scanning methods on timsTOF or if searching with huge libraries) and require peptidoform confidence, an empirical library can indeed be generated using the Ultra-fast mode if you wish to obtain it quicker, we recommend trying this on Orbitrap, Orbitrap Astral and timsTOF data.
This sections focuses on the ways to handle large experiments wherein raw data is being gradually added over a long period of time.
Fast reanalysis. DIA-NN supports adding runs to the experiment and analysing them quickly, without having to reanalyse the whole experiment. For this, first create an empirical spectral library as recommended in the Speed optimisation section above. Analyse each incoming batch of runs separately with this library, make sure that the mass accuracies and the scan window are fixed to specific values. If using QuantUMS, train it on a subset of runs as described in Quantification. Analyse all the runs acquired so far while specifying Reuse .quant files - this will only perform the cross-run steps of the analysis.
Adding runs without changing any quantities. If this is a requirement, we recommend, in addition to the above, to use the Legacy (direct) quantification mode as well as pass --no-maxlfq and --export-quant to DIA-NN. You then obtain raw fragment quantities (as well as raw MS1 quantities that are present regardless of the above settings). Below are some recommendations for obtaining the best quantitative performance (requires R or Python) that roughly correspond to the quantification apporach used by DIA-NN in legacy mode.
-
When you generate your empirical library, select top 3 fragments for each precursor, based on their average scores (also reported when using --export-quant) across the respective (reference) runs. Further, when generating a library, select a set of least variable precursors (e.g. aim to select 40% of all precursors) and calculate their average log2 quantities.
-
When processing DIA-NN output for all runs, calculate non-normalised precursor quantities by summing the selected fragments, replace zero quantities with NA values (important: always use the same logarithm base throughout the script, it's very easy to get incorrect results by accidentally mixing natural logarithms with log2).
-
For each run, calculate the log2 fold-change for each detected precursor with respect to the average log2 levels as recorded across reference runs. Split the retention time range in bins (e.g. 100 bins, aim for 200+ precursors detected per bin). For each bin, calculate either the median or the half sample mode (hsm() function of the modeest package) of those log2 fold-changes of the selected precursors that fall within the bin or the adjacent bins, this value is the normalisation factor for the bin. Interpolate the normalisation factor, to calculate it for every RT. Apply this factor to all precursor identifications depending on their RT. Repeat the procedure with 2x-4x less bins (i.e. larger bins), eventually obtaining 'normalised' precursor quantities.
-
For each batch (here 'batch' refers to any factor that may introduce technical batch effects, e.g. 384 plate used to grow cells), for each precursor, determine a batch correction factor to be applied to its normalised values. For example, if in reference runs a particular precursor was showing 2x higher intensity in HeLa QC injections than in the batch under consideration, then the batch correction factor for this precursor would be equal to 2.0. Alternatively, can calculate batch correction factors also based on average (log-scale) levels of a particular precursor across the biological samples of interest, provided those are appropriately randomised. Apply these correction factors to the normalised precursor quantities.
-
Use the top 1 method to quantify proteins, i.e. in each run the normalised protein quantity is by definition the maximum of the normalised quantities of matching precursors detected at <= 0.01 q-value in this run. May apply protein-level normalisation on top of these, similarly to the precursor-level normalisation, except there is no need to perform RT-binning.
This section is a short introduction to DIA proteomics data analysis without assuming any previous background in proteomics.
Raw data. Each raw DIA acquisition is a collection of MS1 and MS2 (also called MS/MS) spectra. Typically, a single MS1 spectrum and multiple MS2 spectra are recorded in each DIA cycle, and the duration of the cycle (0.3s - 3s) is such as to allow for multiple DIA cycles during the elution of a typical peptide from the LC system. Each MS1 spectrum represents the m/z values (mass over charge) and signal intensity values for the ions generated by the ion source ('precursor' ions), whereas each MS2 spectrum comprises m/z values for their fragments, generated in the collision cells of the mass spectrometer. Typically, to reduce the complexity of MS2 spectra, a mass filter (usually called 'Q1 quadrupole') is used to isolate a particular mass range of precursor ions for fragmentation, e.g. 500-520 m/z or 500.5-501.5 m/z - called 'mass isolation window' or 'selection window' (typically 2 m/z - 50 m/z in DIA).
Spectral libraries. In order to quantify peptides and proteins from the raw data, DIA-NN needs to know which peptides to look for. For example, DIA-NN can be provided with a sequence database (e.g. a reference UniProt proteome in uncompressed .fasta format) as input. DIA-NN can then generate 'precursor ion queries' based on the sequence database. That is, DIA-NN in silico digests the database using the provided enzyme specificity (e.g. trypsin), applies fixed (always present) and variable (may or may not be present) modifications to the resulting peptides and generates 'precursors' as peptides at a particular charge state. Now, given this set of precursors, it's possible to generate all theoretical fragment ions (which are peptide N-terminal and C-terminal fragments produced by the breakage at the peptide bond) and search the raw data for occurances of those. However, raw data search turns out to be much more efficient if the theoretical properties of individual peptides/precursors are predicted with deep learning, i.e. the retention time (RT; term used to refer to elution time of the peptide from the liquid chromatography (LC) system), the ion mobility (IM) and the fragmentation pattern. DIA-NN can do this, with the result being an in silico predicted 'spectral library'. In general, the term 'spectral library' refers to a set of known spectra, retention times and potentially also ion mobility values for selected precursor ions.
Spectral libraries can differ based on how they are generated. What is described above is a predicted spectral library, which may contain millions of entries (e.g. a spectral library based on human UniProt proteome tryptic digest contains about 5 million precursors with charges 1 to 4). Further, spectral libraries can be empirically generated, i.e. contain only precursors observed in a particular experiment. A common strategy has been to perform offline fractionation of a peptide sample (e.g. whole-cell tryptic digest) with subsequent analysis of each of the fractions by LC-MS and the generation of a spectral library comprising the set of confidently identified precursors. This has been traditionally done with DDA, but also works with DIA. In fact, DIA-NN is capable of generating a library from the analysis of any DIA data. That is, one can take a predicted library, search some raw data with it and obtain as a result a much smaller empirical DIA-based library. This library can then be used for a quantitative analysis of the same DIA experiment but also other DIA experiments. The present guide contains detailed explanations of possible workflows based on DIA-NN and guidance on their use.
FDR control. DIA data analysis produces a list of precursors and proteins identified in each of the samples of the experiment. Here 'identified' means that the software expects a particular proportion of those identifications, e.g. 1%, to be false, while the rest, e.g. 99%, are expected to be true. The way DIA-NN does this is by creating a list of likely PSMs (precursor-spectrum matches) and then narrowing it down to only retain PSMs passing certain quality thresholds. This kind of confidence in PSMs is represented by so-called q-values, i.e. 0.01 q-value corresponds to 1% FDR (false-discovery rate). Here it is important to emphasise that if DIA-NN does not report a particular precursor or protein as identified, it does not mean that it is necessarily missing from the sample. Rather this only means that the respective precursor or protein is likely to be relatively low-abundant.
DIA-NN supports both run-specific and global FDR control. Unlike run-specific q-values, global q-values allow to achieve confidence that a particular precursor or protein was correctly identified in at least one mass spectrometry run of the experiment. This is important in particular for large experiments. Indeed, if with each run in the experiment 1% of the identifications are just random noise (at 0.01 run-specific q-value), and all precursors in the library (e.g. 5 million) are equally likely to be falsely identified, then with a large enough experiment all library precursors will eventually be reported as identified in at least one run. Filtering based on global q-values allows to make sure that this does not happen. It also allows DIA-NN to generate high-quality empirical spectral libraries from arbitrary large DIA experiments.
Quantification. Each identified precursor or protein can be quantified in DIA, as all the signals recorded in DIA are quantitative. Here 'quantification' refers to 'relative quantification', wherein the levels of a precursor or a protein can be compared between different runs of the experiment, and hence also between different biological conditions of interest. Importantly, the reported quantities are not expected to be comparable between distinct precursors or between distinct proteins. This limitation originates from the fact that ionisation efficiency of different peptides can vary by orders of magnitude, depending on the peptide sequence. That being said, 'absolute quantification' is possible with DIA. Same as in SRM/PRM, isotopically-labelled peptides can be spiked into the sample in known molar amounts, to quantify their endogenous unlabelled counterparts, using the fact that the ionisation efficiency does not depend on the isotopic composition. Alternatively, absolute quantities of proteins can be estimated with approaches such as IBAQ, although such estimates tend to be of fairly low precision.
Of note, by default DIA-NN reports 'normalised' quantities. This means it makes the best effort to correct for any kind of technical variation in the data that affects all precursors at the same time, such as any kind of pipetting imprecision in sample preparation or different amounts of starting sample material. Indeed, consider a situation when one compares the proteomes of two single cells. If one cell is twice larger, then without normalisation almost every protein will appear differentially expressed. However, such a conclusion is of hardly any biological interest, hence the need for normalisation. Nevertheless, DIA-NN can also provide raw non-normalised intensities, if these are needed given a particular experiment design.
Notes on terminology:
- 'peptide' can be used to refer also to precursor ions;
- 'FDR' can be used interchangeably with 'q-value', although these are, strictly-speaking, not equivalent;
- the term 'library-free analysis' usually refers to using DIA-NN with a predicted spectral library generated from the whole sequence database, that is without an empirical library.
Use of R or Python. We have designed DIA-NN in such a way that the whole workflow requires zero bioinformatics expertise, i.e. indeed all downstream statistical analyses and visualisation can be carried out using MS Excel or similar software as well as dedicated visual tools for proteomics data analysis. However, once you have analysed your data and are now preparing a publication, we do recommend learning some basics of data analysis in R. R is a 'programming language' which nevertheless is very easy to master as coding data processing in R is in a sense similar to formulating a series of bullet points in natural language. Based on experience of a number of colleagues who have had zero prior programming experience, it takes between a day and a week to master R to a level which allows to confidently work with proteomics data (you only need very basics). Indeed, it may take one an hour or so to make several plots in Excel, and only several minutes in R. But most importantly, if you need to adjust your plots or the underlying data, it may be again just minutes in R (change a couple of lines of code and re-run the script), overall it greatly simplifies any kind of work with proteomics data and eventually saves a lot of time. We recommend starting here https://education.rstudio.com/learn/beginner/. This particular section https://r4ds.had.co.nz/data-visualisation.html nicely demonstrates how easy it is to create informative diagrams in R with just one or two lines of code.
Input
- Convert to .dia Convert the selected raw files to DIA-NN's .dia format, for faster subsequent processing, and save them either to the same folder as the respective source raw files or to Temp/.dia dir (Output pane), if the latter is specified. Conversion is recommended for Sciex files, typically makes little difference for Thermo files and is not recommended for Bruker files.
- Spectral library specify the spectral library.
- Add FASTA add one or more FASTA files. FASTA files must always be specified if Protein inference is not disabled (see below) or you would like to obtain protein-matched phosphosite information. The only reason not to specify FASTA files is a situation when you are using an empirical library generated with the use of a different FASTA database that is not available, and in such case specifying FASTA files will overwrite the annotation of protein sequence IDs in the library, potentially replacing protein names, genes and descriptions.
- Reannotate option allows to reannotate the spectral library with protein information from the FASTA database, using the specified digest specificity.
- Contaminants Adds common contaminants from the Cambridge Centre for Proteomics (CCP) database and automatically excludes them from quantification, see the description of the --cont-quant-exclude option. This option applies when generating a predicted spectral library from a FASTA database or analysing using such a library, if it was generated with Contaminants enabled.
Precursor ion generation
- FASTA digest instructs DIA-NN to in silico digest the sequence database and apply specified modifications (see below), to generate a predicted spectral library in silico. This option must be unchecked when analysing raw data.
- Deep learning-based spectra, RTs and IMs prediction instructs DIA-NN to perform deep learning-based prediction of spectra, retention times and ion mobility values. This allows not only to make in silico spectral libraries from sequence databases, but also to replace spectra/RTs/IMs in existing libraries with predicted values. This option must not be used together with raw data analysis, i.e. in silico predicted library generation must be done in a separate step.
- Protease use pre-configured digest specificity, can be customised using the --cut command. Only affects FASTA digest or reannotation.
- Missed cleavages set the maximum number of missed cleavages allowed. Only affects FASTA digest. Increases the search space and hence the analysis time, setting to 1 is almost always the best, in some cases 2 may be beneficial, higher values are likely detrimental.
- Maximum number of variable modifications sets the maximum total number of occupied variable modification sites per peptide, only affects FASTA digest. Reasonable values are usually in 1 to 3 range, setting to 4 or higher is almost always detrimental. Substantially increases the search space and hence the analysis time and RAM usage, may have some detrimental effects on the identification numbers if set too high. Only specify variable modifications if (i) a substantial proportion of detectable peptides is expected to bear the specified number of variable modifications or (ii) you are interested in the biology of a specific modification.
- N-term M excision consider the removal of the protein N-term methionine as a variable modification. Always use this when the database contains actual proteins, rather than just a set of individual peptides.
- C carbimidomethylation declares C(cam) as a fixed modification, see --fixed-mod.
- Ox(M), Ac(N-term), Phospho, K-GG - these options declare the respective modifications as variable, see --var-mod. In case of Phospho declaration applies to S, T and Y. In case of K-GG, instructs DIA-NN to not add diglycine on C-terminal lysine of non-protein-C-term peptides, as enzymes such as trypsin or LysC would not cut after a lysine with a diglycine adduct, see --no-cut-after-mod.
- Precursor and fragment ranges allow to limit which precursors and fragments can be included in the predicted spectral library during FASTA digest. Important: always set minimum precursor charge to 2 when generating predicted libraries for searching timsTOF data, unless the acquisition scheme specifically selects charge 1 precursors for fragmentation (only the case for certain applications, such as immunopeptidomics).
Output
- Reuse .quant files reuse IDs/quantification information from a previous analysis, see Output
- Quantities matrices generate text tables with quantities, see Output
- XICs generate extracted ion chromatograms for DIA-NN Viewer, see Visualisation
- Temp/.dia dir specify where .quant files or converted .dia files will be saved, see Output
- Generate spectral library generate either a predicted library (library name specified by Predicted library below), if no raw data specified, or an empirical DIA-based library (library name specified by Output library below), if analysing raw data.
- Generate Prosit input when performing FASTA digest, create an output file that can be used as input for the Prosit tool. This is helful if you are interested in testing Prosit, however not needed for DIA-NN itself.
- Threads suggest the number of CPU threads for DIA-NN to use. DIA-NN may not scale well beyond 64 threads.
- Precursor FDR (%) set the precursor q-value filtering to be applied to the main output report and the empirical spectral library to be generated. Normally should be kept at 1%, increasing up to 5% may be be helpful if the identification numbers are very low. Increasing beyond 5% is not recommended. Decreasing below 1% may result in worse protein quantification and is also not recommended.
- PDF generate a PDF report with some QC metrics based on the experiment.
Algorithm
-
Mass Accuracy set the MS2 mass tolerance (in ppm), see Changing default settings. Note: there is a view that setting this to very low values may improve quantification. This is not correct, setting to values that are too low may in fact result in a significant loss of quantitative precision.
-
Mass Accuracy MS1 set the MS1 mass tolerance (in ppm), see Changing default settings.
-
Scan window sets the scan window radius to a specific value. Ideally, should be approximately equal to the average number of data points per peak, see Changing default settings.
-
MBR activates a mode in DIA-NN which we call 'match-between-runs'. Note that the algorithm is fundamentally different from MBR in some DDA software. In MBR mode, DIA-NN does two passes over the data. During the first pass, it creates an empirical spectral library from the data. During the second pass, it reanalyses the experiment with this empirical library, which may result in much improved identification numbers, data completeness and quantification accuracy. Always use MBR for quantitative analyses (as opposed to analyses just aimed at creating an empirical library), unless you are analysing with a DIA-based empirical library (in that case do not use MBR). If you are analysing with a project-specific DDA-based library, it may make sense to compare results with and without MBR. For this, analyse first with MBR. You will get both the final MBR report as well as a report based on the first MBR pass (named main_report_name-first-pass.parquet). Compare the identification numbers and data completeness in the two reports. If you decide the performance is higher for the first pass, just uncheck MBR, select Reuse .quant files, change the main output file name and click Run.
MBR is great for small-scale and medium-scale experiments. However, in case of large-scale experiments, one does not need the whole dataset to create a high-quality empirical library. In such cases, we recommend selecting 20 to 100 high quality runs (often selecting just the largest files works well) and create an empirical library from those (that being said, it is perfectly fine to create a library from just a single run as well as thousands of runs). You can then use this library for the analysis of any experiments with matching sample types. Another scenario where one may want to create an empirical library in a separate step is when analysing single-cell data. In this case, it is a good idea to create a library based on some bulk sample runs or runs acquired based on 4-20 cells per sample. Finally, creating an empirical library in a separate step just offers more flexibility. For example, one can then quickly try different quantification modes using Reuse .quant files. Note: do not use empirical libraries generated based on non-timsTOF experiments to analyse timsTOF data. The opposite is fine. -
Unrelated runs determine mass accuracies and scan window, if automatic, independently for different runs, see Changing default settings.
-
Protein inference whether or not to 'infer' proteins present in the sample using a maximum parsimony algorithm. When set to "Off", protein groups from the spectral library are used - this makes sense if protein inference has already been performed during library generation. In all other cases keep protein inference enabled. Protein inference basically means that DIA-NN creates a 'protein group' for each precursor, which is a subset of all protein IDs matched to the precursor. DIA-NN does so aiming to (i) reduce as match as possible the list of proteins matched to the precursor, using any hint it can to prioritise proteins that are more likely to explain the precursor while (ii) ensuring that if two identified precursors are matched to distinct protein groups, then these protein groups do not contain shared proteins. Note that maximum parsimony approaches for protein grouping can result in the following. If precursor X is annotated with proteins A and B, Y - B and C, Z - A and C, then the restriction (ii) will inevitably lead to a situation when protein groups for some of the precursors are missing proteins that could be present in the sample, e.g. groups could be A for X, and C for both Y and Z, whereas protein B is not included in any group. Therefore, the best practice is usually to either (i) use protein groups for any kind of system-scale analysis but manually confirm the differential regulation of proteotypic (i.e. specific to the protein) peptides for specific proteins of particular interest or (ii) rely only on proteotypic peptides.
-
Scoring specifies the type of identification confidence. DIA-NN assigns q-values to target precursors based on how well the respective PSMs score in comparison to in silico generated decoy precursors, i.e. 'faux' peptides not expected to be present in the sample generated by DIA-NN via shuffling or mutation of residues within target peptides, with the respective adjustment of the precursor and fragment masses. The way these decoy peptides are generated is the fundamental factor behind the interpretation of q-values obtained. Please also see Custom decoys.
In the Generic mode, DIA-NN rearranges part of the peptide sequence, aiming to change almost all of the theoretical fragment masses. Since there are no amino acid mutations generated, the precursor mass stays the same (there may be some rare exceptions when DIA-NN cannot sufficiently shuffle the sequence and uses amino acid mutations also in this mode). This is how DIA proteomic data analysis has been traditionally performed: this strategy maximises the numbers of identified proteins. The q-values in this case reflect the confidence that a precursor was correctly matched to a spectrum that has either originated from the same ion species or from an ion species that shares a signficant number of fragment ions (this is a very complex subject, here 'significant' cannot be defined precisely), has the m/z value that falls within the same DIA window, similar enough retention time and (if applicable) similar enough ion mobility. This kind of confidence is sufficient for most experiments of the 'typical proteomics experiment' type. We therefore recommend using this mode if you are interested in protein-level quantification and do not need to confidently distinguish between paralogues or orthologues.
In the Proteoforms mode (new in DIA-NN 2.0), DIA-NN generates decoys by mutating a single residue within each target precursor. This usually results in decoys that share a number of fragment ion masses with the respective targets but differ in the precursor mass. This decoy model is perfect for achieving confidence in the correct assignment of PTMs and correct identification of amino acid substitutions. The resulting q-values can be interpreted as confidence that the spectrum to which a precursor is matched has originated from an ion that not just shares a signficant number of fragment ion masses with it, but also has the same m/z value (within the applicable mass tolerances). Note the two limitations of this strategy (these apply to DDA data too). First, it is not effective in correctly identifying residue localisation within the peptide: phosphate localisation, for example, requires a separate scoring module in DIA-NN. Second, if, for example, the variable modification of interest has the mass equal to several heavy carbon mass deltas (i.e. = n * 1.003355, where n = 1,2,3,4), then the chance of misidnetifying those peptides will be increased even in the Proteoforms mode. This is the reason why DIA-NN has a separate scoring module for channel-specific confidence in plexDIA data analysis. In general, the Proteoforms mode is recommended for (i) most peptidomics applications, (ii) all applications where it's important to distinguish between close paralogues or orthologues and (iii) any data wherein the difference between identification numbers in Generic and Proteoforms modes is negligible, which is the case for many Orbitrap Astral and timsTOF datasets, especially those acquired with longer gradients.
In the Peptidoforms mode, DIA-NN's main q-values correspond to the regular decoy model, same as in the Generic mode, while extra peptidoform q-values provide peptidoform confidence levels. This mode is recommended if (i) you need global (i.e. experiment-wide) peptidoform confidence for some peptides but would also like to maximise the numbers of detected proteins, and (ii) the Proteoforms mode results in a non-negligible reduction of protein identification numbers on the dataset of interest. Note that since two types of q-value filtering are applied simultaneously in Peptidoforms mode, the numbers of precursors with peptidoforms q-values below 0.01 will typically be slightly lower than the numbers of precursors with q-values below 0.01 in the Proteoforms mode (for the first MBR pass), while the actual peptidoform FDR will also be slightly lower. The above differences are typically negligible.
Note: given that protein inference and protein q-value calculation are performed using all identified precursors, it is only the Proteoforms mode that can be used to reliably distinguish paralogue/orthologue proteins originating due to amino acid substitutions, hence the name for this mode. -
Machine learning determines how DIA-NN uses neural networks. The default 'NNs (cross-validated)' mode is recommended for almost all applications, below are some highly technical details explaining it. In general, any kind of ML model is best trained to the extent as to achieve 'mild overfitting' on the training data, as this typically results in best performance. In the 'cross-validated' mode, DIA-NN trains multiple neural networks in such a way that each network is trained (for 4 epochs) only on a subset of the PSMs. Each network is then only used to score PSMs that are neither in its training set nor are sufficiently similar (e.g. same spectrum) to anything in the training set, eliminating any effects overfitting can potentially have on q-value calculation. The 'NN (fast)' mode, in contrast, trains all neural networks on the whole set of PSMs, but just for a single epoch. Of note, the way DIA-NN selects PSMs for training ensures low similarity between those. In combination with single-epoch training, this minimises (but not eliminates) any effects overfitting can have on q-value calculation. However, training for just one epoch results in lower discriminatory power of the networks, ultimately leading to a slightly lower overall identification performance, i.e. lower number of identifications at a given externally controlled FDR. Typically, either difference is negligible. The 'NN (fast)' mode is also considerably faster: for runs with ~100k detectable precursors it takes just several seconds on a modern desktop, as opposed to 15s-30s for the cross-validated mode. We therefore recommend to use 'NNs (fast)' in the following cases: (i) DIA-NN struggles to identify a meaningful number of precursors (with regular/peptidoform or channel-specific q-values) from some runs and you would like to specifically boost identification numbers while slighly relaxing the FDR control in those challenging runs or (ii) you would like to process a large dataset using an empirical library quicker.
-
Proteotypicity this setting affects the proteotypicity definition, calculation of 'protein q-values' and protein inference. We recommend keeping this default (Genes), unless your FASTA does not contain gene information (e.g. UniProt FASTAs do) or you are interested in isoform-level analysis - in that case set this option to Isoform IDs and use --ids-to-names. The proteotypicity definition specified by this option takes effect if DIA-NN infers proteotypicity itself as opposed to extracting this information from the library. This happens if either (i) the spectral library does not contain a column that specifies proteotypicity or (ii) the 'Reannotate' option is used or protein information is absent in the library. In this case, precursors matched only to a single (unique) protein 'entity', as defined by this option (an isoform ID = sequence ID, protein 'name' in the UniProt sense or the gene name), are considered proteotypic. If no FASTA files are provided, DIA-NN acts as if this option was set to 'Isoform IDs'. Whether a precursor is considered proteotypic determines whether it's used to generate the Protein.Q.Value and Genes.MaxLFQ.Unique main report columns, it further may affect protein inference (giving priority to proteins that have more proteotypic precursors identified). This option further has a separate (from specifying proteotypicity) effect on protein q-value calculation, as it defines what is considered a 'protein' for the purposes of this calculation. Further, it affects protein inference: proteins are prioritised also depending on whether they have correct annotation, e.g. in case this option is set to Genes, whether a protein has gene annotation. The 'Genes (species-specific)' option causes DIA-NN to append a species suffix to each gene name when processing a FASTA database, with the suffix extracted from the UniProt-format protein name.
-
Quantification strategy QuantUMS (high-precision) is recommended for most scenarious, use QuantUMS (high-accuracy) for experiments where elimination of any ratio compression bias is critical. Do not use QuantUMS (but instead use the Legacy (direct) mode) if (i) you would like to use Top-N protein quantities - normally this is not recommended, or (ii) you would like to use phosphosite 90 and 99 matrices generated by DIA-NN, or (iii) you are benchmarking/optimising the LC-MS method, or (iv) you would like to estimate the absolute quantities using IBAQ or a similar approach. Note that if you analyse with an empirical library without MBR, you can quickly switch from QuantUMS to Legacy quantification or vice versa by checking Reuse .quant files, changing the main output report name and clicking Run. Please see Quantification for a detailed guide on different types of quantification information provided by DIA-NN and some best practices for statistical analysis of proteomics data.
-
Cross-run normalisation whether to use global, RT-dependent (recommended) or also signal-dependent (experimental, be very careful about it) cross-run normalisation. Normalisation can also be disabled completely. Please see Quantification for a detailed guide.
-
Library generation this setting determines if and how empirical RTs/IMs and spectra are added to the newly generated library, instead of the theoretical values. IDs, RT & IM profiling is strongly recommended for almost all workflows. When analysing with a high-quality project-specific library, can switch to IDs profiling. Full profiling means always using empirical information, and might only be beneficial (in very rare cases) when having less than ~1000 peptides identified per run.
-
Speed and RAM usage this setting is primarily useful for library-free analyses. The first three modes will typically have little difference in terms of ID numbers, while the Ultra-fast mode is rather extreme: up to 5x faster, but ID numbers are not as good. The setting affects only the first pass when using MBR. Therefore, in many cases, analysing in Ultra-fast mode with large predicted libraries with MBR enabled may produce same identification numbers during the second MBR pass, especially if the data has been acquired on Orbitrap, Orbitrap Astral or timsTOF instruments.
Description of available options/commands
Note that some options below are strongly detrimental to performance and are only there for benchmarking purposes. So the recommendation is to only use the options which are expected to be beneficial for a particular experiment (e.g. those recommended in the present guide) based on some clear rationale.
- --cal [N] set the calibration mode, valid values are 0 (off), 1, 3, 5 and 7, default is 7
- --cfg [file name] specifies a file to load options/commands from
- --channel-run-norm normalisation of multiplexed samples will be performed in run-specific manner, i.e. to perform normalisation, for each precursor ion DIA-NN will sum the respective channels within each run and will normalise these sums across runs: use e.g. for protein turnover SILAC experiments
- --channel-spec-norm normalisation of multiplexed samples will be performed in channel-specific manner, i.e. each channel in each run is treated as a separate sample to be normalised: use to analyse experiments wherein multiplexing of independent samples is used to boost throughput
- --channels [channel 1]; [channel 2]; ... lists multiplexing channels, wherein each channel declaration has the form [channel] = [label group],[channel name],[sites],[mass1:mass2:...], wherein [sites] has the same syntax as for --var-mod and if N sites are listed, N masses are listed at the end of the channel declaration. The spectral library will be automatically split into multiple channels, for precursors bearing the [label group] modification. To add the latter to a label-free spectral library, can use --lib-fixed-mod, e.g. --fixed-mod SILAC,0.0,KR,label --lib-fixed-mod SILAC. See Multiplexing using plexDIA for usage examples. Currently, [label group] must be the same for all channels and the channels must be listed in such an order that the channel mass shifts are non-decreasing for each modification site.
- --clear-mods makes DIA-NN 'forget' all built-in modification (PTM) names
- --cont-quant-exclude [tag] peptides corresponding to protein sequence ids tagged with the specified tag will be excluded from normalisation as well as quantification of protein groups that do not include proteins with the tag
- --convert makes DIA-NN convert the mass spec files to the .dia format. The files are either saved to the same location as the input files, or in the Temp/.dia dir, if it is specified (in the GUI or using the --temp option)
- --cut [specificty 1],[specificity 2],... specifies cleavage specificity for the in silico digest. Cleavage sites (pairs of amino acids) are listed separated by commas, '*' indicates any amino acid, and '!' indicates that the respective site will not be cleaved. Examples: "--cut K*,R*,!*P" - canonical tryptic specificity, "--cut " - digest disabled, "--cut ** --missed-cleavages 100" will make DIA-NN perform a non-specific digest (first do this for a single peptide length, to estimate the numbers of precursors generated)
- --decoy-channel [channel] specifies the decoy channel masses, wherein [channel] has the same syntax as for --channels
- --dg-keep-nterm [N] do not change the first N residues when generating decoys, default N = 1
- --dg-keep-cterm [N] do not change the last N residues when generating decoys, default N = 1
- --dg-min-shuffle [X] aim for any fragment mass shift introduced by shuffling during decoy generation to exceed X in absolute value, default 5.0
- --dg-min-mut [X] aim for the precursor mass shift during mutation during decoy generation to be at least X in absolute value, default 15.0
- --dg-max-mut [X] aim for the precursor mass shift during mutation during decoy generation not to exceed X in absolute value, default 50.0
- --dir [folder] specifies a folder containing raw files to be processed. All files in the folder must be in .raw, .wiff, .mzML or .dia format
- --dir-all [folder] as --dir, but recursive over subfolders
- --direct-quant disable QuantUMS and use legacy DIA-NN quantification algorithms instead, also disables channel-specific protein quantification when analysing multiplexed samples
- --dl-no-fr when using the deep learning predictor, prediction of fragment intensities will not be performed
- --dl-no-im when using the deep learning predictor, prediction of ion mobilities will not be performed
- --dl-no-rt when using the deep learning predictor, prediction of retention times will not be performed
- --duplicate-proteins instructs DIA-NN not to skip entries in the sequence database with duplicate IDs (while by default if several entries have the same protein ID, all but the first entry will be skipped)
- --export-quant add fragment quantities, fragment IDs and quality information to the .parquet output report
- --ext [string] adds a string to the end of each file name (specified with --f)
- --f [file name] specifies a run to be analysed, use multiple --f commands to specify multiple runs
- --fasta [file name] specifies a sequence database in FASTA format (full support for UniProt proteomes), use multiple --fasta commands to specify multiple databases
- --fasta-filter [file name] only consider peptides matching the stripped sequences specified in the text file provided (one sequence per line), when processing a sequence database
- --fasta-search instructs DIA-NN to perform an in silico digest of the sequence database
- --fixed-mod [name],[mass],[sites],[optional: 'label'] - adds the modification name to the list of recognised names and specifies the modification as fixed. Same syntax as for --var-mod. Has an effect of (i) applying fixed modifications during FASTA digest or (ii) declaring fixed modifications that can be applied to a library with --lib-fixed-mod
- --force-swissprot only consider SwissProt (i.e. marked with '>sp|') sequences when processing a sequence database
- --full-profiling enable using empirical spectra for empirical library generation
- --full-unimod loads the complete UniMod modification database and disables the automatic conversion of modification names to the UniMod format
- --gen-spec-lib instructs DIA-NN to generate a spectral library
- --gen-fr-restriction annotates the library with fragment exclusion information, based on the runs being analysed (fragments least affected by interferences are selected for quantification, why the rest are excluded)
- --global-norm instructs DIA-NN to use simple global normalisation instead of RT-dependent normalisation
- --high-acc QuantUMS settings will be otimised for maximum accuracy, i.e. to minimise any ratio compression quantitative bias
- --ids-to-names protein sequence ids will also be used as protein names and genes, any information on actual protein names or genes will be ignored
- --id-profiling set the empirical library generation mode to IDs profiling
- --il-eq when using the 'Reannotate' function, peptides will be matched to proteins while considering isoleucine and leucine equivalent
- --im-acc [X] a parameter that should roughly correspond to the magnitude of observed IM value differences between different ion species matching the same precursor, default X = 0.02
- --im-model [file] specifies the file containing the IM deep learning prediction model
- --im-prec [X] a parameter that should roughly correspond to the magnitude of IM errors due to noise, default X = 0.01
- --im-window [x] IM extraction window will not be less than the specified value
- --individual-mass-acc mass accuracies, if set to automatic, will be determined independently for different runs
- --individual-reports a separate output report will be created for each run
- --individual-windows scan window, if set to automatic, will be determined independently for different runs
- --int-removal 0 disables the removal of interfering precursors, not recommended
- --lib [file] specifies a spectral library. The use of multiple --lib commands (experimental) allows to load multiple libraries in .tsv format
- --lib-fixed-mod [name] in silico applies a modification, previously declared using --fixed-mod, to a spectral library
- --mass-acc [N] sets the MS2 mass accuracy to N ppm
- --mass-acc-cal [N] sets the mass accuracy used during the calibration phase of the search to N ppm (default is 100 ppm, which is adjusted automatically to lower values based on the data)
- --mass-acc-ms1 [N] sets the MS1 mass accuracy to N ppm
- --matrices output quantities matrices
- --matrix-qvalue [x] sets the q-value used to filter the output matrices
- --matrix-spec-q [x] run-specific protein q-value filtering will be used, in addition to the global q-value filtering, when saving protein matrices. The ability to filter based on run-specific protein q-values, which allows to generate highly reliable data, is one of the advantages of DIA-NN
- --max-pep-len [N] sets the maximum precursor length for the in silico library generation or library-free search
- --max-pr-charge [N] sets the maximum precursor charge for the in silico library generation or library-free search
- --mbr-fix-settings when using the 'Unrelated runs' option in combination with MBR, the same settings will be used to process all runs during the second MBR pass
- --met-excision enables protein N-term methionine excision as variable modification for the in silico digest
- --min-cal [N] provide a guidance to DIA-NN suggesting the minimum number of IDs to use for mass calibration
- --min-class [N] provide a guidance to DIA-NN suggesting the minimum number of IDs to use for linear classifier training
- --min-corr [X] forces DIA-NN to only consider peak group candidates with correlation scores at least X
- --min-fr specifies the minimum number of fragments per precursors in the spectral library being saved
- --min-peak sets the minimum peak height to consider. Must be 0.01 or greater
- --min-pep-len [N] sets the minimum precursor length for the in silico library generation or library-free search
- --min-pr-charge [N] sets the minimum precursor charge for the in silico library generation or library-free search
- --min-pr-mz [N] sets the minimum precursor m/z for the in silico library generation or library-free search
- --missed-cleavages [N] sets the maximum number of missed cleavages
- --mod [name],[mass],[optional: 'label'] declares a modification name. Examples: "--mod UniMod:5,43.005814", "--mod SILAC-Lys8,8.014199,label"
- --no-batch-mode disable batch mode, consequently, use all precursors for calibration
- --no-calibration disables mass calibration, not recommended
- --no-cut-after-mod [name] discard peptides generated via in silico cuts after residues bearing a particular modification
- --no-decoy-channel disables the use of a decoy channel for channel q-value calculation
- --no-fragmentation DIA-NN will not consider fragments other than included in the spectral library
- --no-fr-selection the selection of fragments for quantification based on the quality assessment of the respective extracted chromatograms will be disabled
- --no-isotopes do not extract chromatograms for heavy isotopologues
- --no-lib-filter the input library will be used 'as is' without discarding fragments that might be harmful for the analysis; use with caution
- --no-maxlfq disables MaxLFQ for protein quantification
- --no-ms1 do not consider MS1 data, use only during method optimisation to evaluate the impact of MS1 on the data quality
- --no-norm disables cross-run normalisation
- --no-peptidoforms disables automatic activation of peptidoform scoring when variable modifications are declared, not recommended
- --no-prot-inf disables protein inference (that is protein grouping) - protein groups from the spectral library will be used instead
- --no-prot-norm disable protein-level normalisation
- --no-quant-files instructs DIA-NN not to save .quant files to disk and store them in memory instead
- --no-rt-norm disable RT-dependent normalisation
- --no-rt-window disables RT-windowed search
- --no-skyline do not generate .skyline.speclib
- --no-stats disables the generation of the stats file
- --no-swissprot instruct DIA-NN not to give preference for SwissProt proteins when inferring protein groups
- --original-mods disables the automatic conversion of known modifications to the UniMod format names known to DIA-NN
- --out [file name] specifies the name of the main output report. The names of all other report files will be derived from this one
- --out-lib [file name] specifies the name of a spectral library to be generated
- --out-lib-copy copies the spectral library used into the output folder
- --peak-boundary [X] if the fragment or MS1 signal decays below its max / X, this is considered the boundary of the elution peak, affects among other algorithms the calculation of start/stop RT values reported in the main .parquet report
- --peak-translation instructs DIA-NN to take advantage of the co-elution of isotopologues, when identifying and quantifying precursors; automatically activated when using --channels
- --peptidoforms enables peptidoform confidence scoring
- --pg-level [N] controls the protein inference mode, with 0 - isoforms, 1 - protein names (as in UniProt), 2 - genes
- --predict-n-frag [N] specifies the maximum number of fragments predicted by the deep learning predictor, default value is 12
- --predictor instructs DIA-NN to perform deep learning-based prediction of spectra, retention times and ion mobility values
- --prefix [string] adds a string at the beginning of each file name (specified with --f) - convenient when working with automatic scripts for the generation of config files
- --prosit export prosit input based on the FASTA digest
- --proteoforms enables the proteoform confidence scoring mode
- --pr-filter [file name] specify a file containing a list of precursors (same format as the Precursor.Id column in DIA-NN output), FASTA digest will be filtered to only include these precursors
- --qvalue [X] specifies the precursor-level q-value filtering threshold
- --quant-acc [X] sets the precision-accuracy balance for QuantUMS to X, where X must be between 0 and 1
- --quant-ori-names .quant files will retain original raw file names even if saved to a separate directory, convenient for .quant file manipulation
- --quant-fr [N] sets the number of top fragment ions among which the fragments that will be used for quantification are chosen for the legacy (pre-QuantUMS) quantification mode. Default value is 6
- --quick-mass-acc (experimental) when choosing the MS2 mass accuracy setting automatically, DIA-NN will use a fast heuristical algorithm instead of IDs number optimisation
- --quant-no-ms1 instructs QuantUMS not to use the recorded MS1 quantities directly
- --quant-params [params] use previously obtained QuantUMS parameters
- --quant-sel-runs [N] instructs QuantUMS to train its parameters on N automatically chosen runs, to speed up training for large experiments, N here must be 6 or greater
- --quant-tims-sum for slice/scanning timsTOF methods, calculate intensities by taking the sum of all cognate signals across the cycle, as opposed to taking the maximum, use this for Synchro-PASEF
- --quant-train-runs [N1]:[N2] instructs QuantUMS to train its parameters on runs with indices in the range N1 to N2 (inclusive), e.g. --quant-train-runs 0:5 will perform training on 6 runs with indices 0 to 5
- --reanalyse enables MBR
- --reannotate reannotate the spectral library with protein information from the FASTA database, using the specified digest specificity
- --ref [file name] (experimental) specify a special (small) spectral library which will be used exclusively for calibration - this function can speed up calibration in library-free searches; this special library must have the same RT and IM scales as the main predicted/experimental library used for the analysis; use with caution
- --regular-swath all runs will be analysed as if they were not Scanning SWATH runs
- --report-decoys save decoy PSMs to the main .parquet report
- --restrict-fr some fragments will not be used for quantification, based on the value in the ExcludeFromAssay spectral library column; marking all fragments of a precursor as excluded will, together with --restrict-fr, suppress the use of this precursor for protein quantification whenever possible
- --rt-model [file] specifies the file containing the RT deep learning prediction model
- --rt-profiling set the empirical library generation mode to IDs, RT and IM profiling
- --rt-window [X] RT extraction window will not be less than the specified value
- --scanning-swath all runs will be analysed as if they were Scanning SWATH runs
- --semi when using the 'Reannotate' function, a peptide will be matched to a protein also if it could be obtained with one specific and one non-specific cut (at either of the termini)
- --sig-norm enable signal-dependent normalisation, not recommended
- --skip-unknown-mods instructs DIA-NN to ignore peptides with modifications that are not supported by the deep learning predictor, when performing the prediction
- --smart-profiling enables an intelligent algorithm which determines how to extract spectra, when creating a spectral library from DIA data, use with --full-profiling. The performance has so far always been observed to be comparable to IDs, RT and IM profiling
- --species-genes instructs DIA-NN to add the organism identifier to the gene names - useful for distinguishing genes from different species, when analysing mixed samples. Works with UniProt sequence databases.
- --species-ids instructs DIA-NN to add the organism identifier to the sequence ids/protein isoform ids - useful for distinguishing protein ids from different species, when analysing mixed samples. Works with UniProt sequence databases.
- --sptxt-acc [N] sets the fragment filtering mass accuracy (in ppm) when reading .sptxt/.msp libraries
- --tag-to-ids [tag] proteins that have the respective FASTA header start with tag (i.e. the string following '>' starts with tag) will have the tag incorporated in the respective protein sequence ids, names and genes
- --temp [folder] specifies the Temp/.dia directory
- --threads [N] specifies the number of CPU threads to use
- --time-corr-only restrict machine learning during calibration
- --time-corr-only-force restrict the use of machine learning for peak group selection during all search stages
- --tims-min-int [N] specifies the minimum peak intensity (an integer ion count) to be considered when loading timsTOF data
- --tims-ms1-cycle [N] merge the MS/MS spectra from N consecutive cycles, each cycle defined as the set of MS/MS scans following an MS1 scan, use this option only for slice/scanning timsTOF data and always test if the performance is better than without it
- --tokens [file] specify the file name for the text file that contains a mapping between modified residues or N-term modifications and integers in the range 0-255, this file is used by the deep learning predictor
- --top [N] set N for the top N protein quantification, default N = 1
- --tune-im tune the IM deep learning predictor, requires --tune-lib
- --tune-lib [file] specifies the library to be used for deep learning predictor fine-tuning, may require declaring unknown modifications with --mod
- --tune-lr [X] specifies fine-tuning learning rate, default is 0.00001
- --tune-restrict-layers keep RNN layer weights fixed when fine-tuning
- --tune-rt tune the RT deep learning predictor, requires --tune-lib
- --use-quant use existing .quant files, if available
- --verbose [N] sets the level of detail of the log. Reasonable values are in the range 0 - 4
- --var-mod [name],[mass],[sites],[optional: 'label'] - adds the modification name to the list of recognised names and specifies the modification as variable. [sites] can contain a list of amino acids and 'n' which codes for the N-terminus of the peptide. '*n' indicates protein N-terminus. Examples: "--var-mod UniMod:21,79.966331,STY" - phosphorylation, "--var-mod UniMod:1,42.010565,*n" - N-terminal protein acetylation. Similar to --mod can be followed by 'label'. Has an effect of (i) applying variable modifications during FASTA digest or (ii) declaring modifications to be localised during raw data analysis
- --var-mods sets the maximum number of variable modifications
- --xic [optional: X] instructs DIA-NN to extract MS1/fragment chromatograms for identified precursors within X seconds from the elution apex, with X set to 10s if not provided; the chromatograms are saved in .parquet files (one per run) located in a folder that is created in the same location as the main output report; equivalent to the 'XICs' option in the GUI
- --xic-theoretical-fr makes DIA-NN extract chromatograms for all theoretical charge 1-2 fragment ions, including those with common neutral losses, if --xic is used
- --window [N] sets the scan window radius to a specific value. Ideally, should be approximately equal to the average number of MS/MS data points per peak
Description of selected columns in the main .parquet report
- Run raw file name without file path
- Channel channel-identifier for the respective precursor, as specified in --channels
- Precursor.Id precursor identifier, i.e. modified peptide sequence with a specific charge, channel-oblivious
- Modified.Sequence modified peptide sequence, channel-oblivious
- Stripped.Sequence amino acid sequence, no modifications
- Precursor.Charge the charge of the precursor
- Precursor.Lib.Index index of the precursor in the internal representation used by DIA-NN for the spectral library
- Decoy 0 or 1 based on whether the precursor is decoy, relevant when using --report-decoys
- Proteotypic 0 or 1 based on whether the precursor is considered proteotypic
- Precursor.Mz library m/z value of the precursor
- Protein.Group inferred proteins. See the description of the Protein inference GUI setting and the --relaxed-prot-inf option.
- Protein.Ids - all proteins matched to the precursor in the library or, in case of library-free search, in the sequence database
- Protein.Names names (UniProt names) of the proteins in the Protein.Group
- Genes gene names corresponding to the proteins in the Protein.Group. Note that protein and gene names will only be present in the output if either (i) Protein inference is disabled in DIA-NN and no sequence (FASTA) database is provided - then protein & gene names from the spectral library will be used, or (ii) protein IDs in the Protein.Group can be found in the sequence database provided, or (iii) the Reannotate function is used. (ii) and (iii) also require that DIA-NN recognises protein & gene names as recorded in the sequence database (e.g. the UniProt format is supported)
- RT the retention time (RT) of the PSM in minutes
- iRT reference RT as recorded in the spectral library
- Predicted.RT predicted RT based on the iRT
- Predicted.iRT predicted iRT based on the RT
- IM, iIM, Predicted.IM, Predicted.iIM - similarly for ion mobility (RT) as for RT
- Precursor.Quantity MS2-based quantity of the precursor (charged peptide), non-normalised
- Precursor.Normalised MS2-based quantity of the precursor (charged peptide), normalised, if normalisation is not disabled
- Ms1.Area non-normalised MS1 peak area
- Ms1.Normalised normalised MS1 peak area
- Ms1.Apex.Area MS1 peak integrated only in the vicinity of the apex
- Ms1.Apex.Mz.Delta difference between observed precursor m/z and the theoretical value
- Normalisation.Factor normalisation factor applied to the precursor in the specific run, i.e. normalised quantity = normalisation factor X non-normalised quantity
- Quantity.Quality when using QuantUMS is equal to 1.0 / (1.0 + SD), where SD is the estimated standard deviation of the LC-MS-derived error in relative precursor quantification
- Empirical.Quality when using QuantUMS reflects the agreement of relative precursor quantities obtained using different quantitative features (MS1 / fragment ions)
- Normalisation.Noise reflects the noisiness of the RT-dependent normalisation factor calculated for the precursor
- Ms1.Profile.Corr a score that reflects the similarity between MS1 and MS2 extracted ion chromatograms (XICs)
- Evidence the sum of correlation scores for the XICs of the top 6 library fragments and the precursor m/z (MS1)
- Mass.Evidence a score that reflects MS2-level evidence for the spectrum originating from a an ion with m/z matching that of the reported precursor
- Channel.Evidence a score that reflects the quality of evidence supporting correct channel assignment for the precursor
- Ms1.Total.Signal.Before total Ms1 signal recorded in the MS1 spectrum immediately preceding the apex MS2 scan of the PSM
- Ms1.Total.Signal.After total Ms1 signal recorded in the MS1 spectrum immediately following the apex MS2 scan of the PSM
- RT.Start and RT.Stop peak boundaries
- FWHM estimated peak width at half-maximum; note that the accuracy of such estimates sometimes strongly depends on the DIA cycle time and sample injection amount, i.e. they can only be used to evaluate chromatographic performance in direct comparisons with similar settings, including the scan window; another caveat is that FWHM does not reflect any peak tailing
- PG.TopN Normalised protein group quantity calculated using top N method (default N = 1, i.e. max normalised precursor quantity in the given run is used), relevant in Legacy (direct) quantification mode
- PG.MaxLFQ QuantUMS or MaxLFQ normalised quantity for the protein group, channel-specific
- Genes.TopN as PG.TopN, but calculated for the gene group
- Genes.MaxLFQ as PG.MaxLFQ, but calculated for the gene group
- Genes.MaxLFQ.Unique as Genes.MaxLFQ, but calculated solely using proteotypic precursors; note the effects of Proteotypicity (GUI option) and --ids-to-names
- PG.MaxLFQ.Quality when using QuantUMS reflects the quality of PG.MaxLFQ
- Genes.MaxLFQ.Quality when using QuantUMS reflects the quality of Genes.MaxLFQ
- Genes.MaxLFQ.Unique.Quality when using QuantUMS reflects the quality of Genes.MaxLFQ.Unique
- Q.Value run-specific precursor q-value
- PEP run-specific posterior error probability for the precursor
- Global.Q.Value global precursor q-value
- Lib.Q.Value q-value for the respective library entry, 'global' if the library was created by DIA-NN. In case of MBR, this applies to the empirical library created based on the first MBR pass
- Peptidoform.Q.Value q-value reflecting peptidoform confidence in Peptidoforms scoring mode
- Global.Peptidoform.Q.Value q-value reflecting global peptidoform confidence in Peptidoforms scoring mode
- Lib.Peptidoform.Q.Value peptidoform q-value for the respective library entry. In case of MBR, this applies to the library created based on the first MBR pass
- PTM.Site.Confidence run-specific confidence in site localisation (=estimated 1 - Site.PEP, where Site.PEP is the posterior error probability for incorrect modification localisation), reported for the leas confident occupied site
- Site.Occupancy.Probabilities modified peptide sequence with estimated variable modification sites occupancy probabilities given in curly brackets
- Protein.Sites list of protein residues corresponding to the occupied modification sites, requires a FASTA database to be provided
- Lib.PTM.Site.Confidence the site localisation confidence recorded in the library. In case of MBR, this applies to the library created based on the first MBR pass
- Translated.Q.Value the q-value a precursor would have had if it was not 'translated' between multiplexed channels
- Channel.Q.Value reflects the confidence that the precursor is indeed present in the respective channel
- PG.Q.Value run-specific q-value for the protein group, channel-specific
- PG.PEP run-specific posterior error probability for the protein group, channel-specific
- GG.Q.Value run-specific q-value for the gene group (identified by the value in the Genes column), channel-specific
- Protein.Q.Value run-specific q-value for the unique protein/gene, that is protein/gene identified with proteotypic (=specific to it) peptides, not channel-specific
- Global.PG.Q.Value global q-value for the protein group
- Lib.PG.Q.Value protein group q-value for the respective library entry, 'global' if the library was created by DIA-NN. In case of MBR, this applies to the empirical library created based on the first MBR pass
Filtering the main report
While the matrix output of DIA-NN is 'ready to use', the main report is designed to be filtered, the idea here is to record any potentially useful information in the main report so that it can be retrieved later on, including low-confidence matches and quantities. The filtering can typically be done in a single line of R or Python code.
The following filters are recommended in most cases:
- Q.Value at 0.01, done by DIA-NN automatically if analysing with Precursor FDR set to 1%
- Lib.Q.Value at 0.01, and in addition Global.Q.Value unless using an empirical library created based on the same samples (e.g. if using MBR filtering based on Global.Q.Value is unnecessary)
- Lib.PG.Q.Value and Global.PG.Q.Value, same as above
- Lib.Peptidoform.Q.Value and Global.Peptidoform.Q.Value, same as above - in Peptidoforms scoring mode
- PG.Q.Value at 0.01 to 0.05, typically 0.05 is fine
- Channel.Q.Value at 0.01 to 0.5 (not a typo) when analysing multplexed (plexDIA) data
Sometimes also good to filter:
- Quantity.Quality at 0.5 or higher when using QuantUMS, usually makes sense to choose a filter threshold that does not substantially reduce identification numbers
- PG.MaxLFQ.Quality at 0.7 or higher when using QuantUMS
- An averaged value of the above for the specific precursor/protein across the experiment
Q: Why DIA-NN?
A: Proteomic depth, quantitative precision, reliability and speed. In particular, DIA-NN is transformative for (i) experiments acquired with fast chromatographic gradients and (ii) library-free analyses. Further, DIA-NN is the basis of such technologies as plexDIA, Slice-PASEF and QuantUMS.
Q: I have a regular experiment, which parts of this guide do I really need to read?
A: Getting Started is sufficient to start working with DIA-NN. Afterwards can also look at Changing default settings. If something is not working, check Raw data formats and Spectral library formats.
Q: I am new to DIA proteomics, what papers would you suggest?
A: Ludwig et al 2018 is an excellent introduction to DIA. The DIA-NN paper describes how neural network classifier in DIA-NN enables confident proteomics with fast methods and is an example of how to benchmark DIA software performance. Please note that the field of DIA proteomics is developing very rapidly, and things get outdated quickly. For example, the latest version of DIA-NN is overwhelmingly better in every aspect than DIA-NN 1.6, which was featured in the initial DIA-NN paper.
Q: How do I ensure reproducibility of my analyses?
A: DIA-NN has a strong focus on reproducibility. First, the pipeline functionality (and the ability to save pipelines) allows to conveniently store the experiment settings - with DIA-NN it can never happen that a user forgets what settings were used for a particular analysis. Second, DIA-NN's log contains complete information on the settings used and is automatically saved for each experiment, along with a separate pipeline step - which allows to easily rerun the analysis, even if the user forgot to save the whole pipeline.
In terms of computational reproducibility, please note the following:
- DIA-NN's analysis using a spectral library will always be the same across different Windows machines.
- In silico prediction of spectra/RTs/IMs using deep learning will yield slightly different results depending on the instruction set supported by the CPU - that's why DIA-NN prints this instruction set upon being launched. This is a consequence of computational optimisations for modern architectures as implemented in the Pytorch deep learning framework used by DIA-NN.
- DIA-NN's analysis on any Linux machine will often be identical but may be slightly different from that on Windows, as certain mathematical functions may be calculated slightly differently by Linux libraries. It is not guaranteed but very likely that analysis results will be the same across all reasonable Linux machines. So for reproducibility purposes in practice it is enough to just keep track of what kind of Linux distribution with what glibc version is being used.
Q: Do I need imputation?
A: For many downstream applications - no. Most statistical tests are fine handling missing values. When imputation is necessary (e.g. for certain kinds of machine learning, or when a protein is completely absent in some of the biological conditions), we prefer to perform it on the protein level. Note that many papers that discuss imputation methods for proteomic data benchmark them on DDA data. The fundamental difference between DDA and DIA is that in DIA if a value is missing it is a lot more likely that the respective precursor/protein is indeed quite low-abundant. Because of this, minimal value imputation, for example, performs better for DIA than it does for DDA. An important consideration with imputation is that Gaussian statistical methods (like t-test) should be used with caution on imputed data, if the latter is strongly non-Gaussian.
Q: Can I obtain the quantities of the individual fragments?
A: You can use the --export-quant option to instruct DIA-NN to save fragment peak heights (non-normalised) to the .parquet report, along with quality scores for the respective extracted elution profiles. This allows to devise custom quantification strategies. Note that in that case you can still leverage the normalisation factor calculated by DIA-NN for each precursor identification.
Q: When designing an experiment, what number of data points per peak should I aim for?
A: We typically aim for 3-3.5 data points per peak at FWHM (full width at half maximum), which is about 6-7 data points in total. If you set Scan window to 0 in DIA-NN, it will print its estimate for the average "Peak width" - this is actually the estimate for the number of data points at FWHM. For example, for LFQbench data this figure is approximately 3.0-3.1. This is enough to achieve good precision of quantification. In terms of identification performance, methods with longer cycle time and hence less data points per peak typically yield better ID numbers, and thus are preferable when the sole purpose is to generate a spectral library, which would then be used on a different dataset.
Q: How useful are MS1 scans for DIA?
A: For what is called 'regular proteomics' - not particularly useful. However, MS1 information is crucially important for distinguishing peptidoforms/PTMs and for multiplexing. Also, MS1 scans are, in most cases, quite inexpensive time-wise, so it is almost always worth having them.
Q: How to choose the FDR/q-value threshold?
A: Depends on the downstream analysis. In general, relaxing FDR filtering up to 5% might be reasonable in some cases. But it is only justified if it leads to a significant improvement in identification numbers. One option is to reduce the numbers of missing values by applying 1% global q-value filter and, say, 3% or 5% run-specific q-value filter. Again, only justified if the numbers of missing values are reduced significantly. Numbers of modified peptides detected sometimes strongly depend on the q-value filter threshold chosen, with 2% or 3% yielding significantly more IDs than 1% filtering. In such cases it might be worth it to also relax global q-value filter to 2% or 3%.
When it comes to protein q-value filtering, it is a common practice to only filter using global 1% q-value, to ensure at least 99% of proteins are correctly detected in at least one run in the experiment. This also guarantees low effective run-specific protein FDR, provided the spectral library used contains a lot more precursors and proteins than are actually detectable in the DIA experiment. For example, if the spectral library contains 11k proteins and 500k precursors, but only 3k proteins pass 1% global q-value filter, and only 100k precursors map to these proteins, then if the output is filtered at 1% run-specific precursor q-value and 1% global protein q-value, the effective run-specific protein FDR will also be well controlled at about 1%. However, if almost all the proteins in the library pass 1% global q-value threshold, 1% global protein q-value filtering has no effect. As a consequence, the effective run-specific protein FDR is only controlled indirectly by run-specific precursor q-value, and empirically it usually results in 5-10% effective run-specific protein FDR, given 1% run-specific precursor q-value filtering.
Is having 5% effective run-specific protein FDR a problem? In many cases it is not. Suppose a protein passes 1% run-specific protein q-value filtering in 50% of the runs, and in the other 50% it is quantified but has run-specific q-value > 1%. What are the options? One is to just use those quantities, most of them are likely to be correct. Another is to impute by replacing them via random sampling from some distribution, for example. For most applications the first option is clearly better.
However in some cases one might want to be very confident in protein identifications in specific runs. And here what helps a lot is the ability of DIA-NN to calculate run-specific protein q-values, which is absent from many alternative software options.
There is an important distinction between q-values and posterior error probabilities (PEPs). The q-values allow to select a subset of identifications while controlling the proportion of false ones. The PEPs reflect the probabilities that individual identifications are correct. It is not uncommon for identifications with q-values below 1% to have PEP values above 50%, making them unreliable for quantification. Therefore, if you require extra confidence in specific identifications, it does make sense to apply a PEP filter, respectively at the precursor (PEP) or protein (PG.PEP) level.
Q: What is normalisation and how does it work?
A: The purpose of normalisation is to cancel out some of the undesirable effects which can occur during sample preparation and acquisition. For example, pipetting errors can result in different total amounts being injected, and thus signals of different intensity being recorded by the mass spec. The basic global normalisation addresses this. It should in general be used under the following assumption: when only considering 'biological' variability of interest, most of the peptides are not differentialy abundant, or the numbers of upregulated / downregulated (in comparison to experiment-wide averages) peptides in each sample are about the same. If this condition is not satisfied (e.g. when analysing fractions obtained with some separation technique, like SEC, for instance), then normalisation should not be used.
DIA-NN also supports a more advanced normalisation method, which is enabled by default: RT-dependent normalisation. Sometimes, undesirable technical effects can lead to some samples having either hydrophilic or hydrophobic peptides depleted, relative to other samples. These effects are corrected by RT-dependent normalisation. Potentially it might also correct for spray stability issues.
The choice of the normalisation strategy in DIA-NN determines the way .Normalised and .MaxLFQ quantities (in the main report), as well as values in output matrices are calculated. So if normalisation is disabled, these quantities are actually non-normalised.
Q: How to compare the identification performance of two DIA software tools?
A: This is often tricky because FDR estimates are never precise. For example, recently we have benchmarked (see Supplementary Figure S3 therein) DIA-NN on data acquired with different chromatographic gradients, and what we observed is that although DIA-NN is always somewhat conservative in its FDR estimates (that is when it reports FDR = 1%, the effective FDR as estimated with a two-species spectral library method is in fact lower than 1%), the accuracy of its estimates varies a bit, depending on the data. When comparing different software tools, the differences in FDR estimation accuracy will often be even larger, due to different algorithms used. Note also that certain kinds of data are very sensitive to the FDR threshold, e.g. the ID number can be, say, 20k peptides at 1% FDR, and go up to 30k at 2%. So if software A says it's 1%, real FDR is also 1%, and it reports 20k, while software B says it's 1%, but real FDR is 2%, and it still reports 20k, these two software tools are actually quite far from each other in terms of real performance.
Because of this, it is important to always independently estimate the FDR using an unbiased benchmarking procedure, which is not affected by the internal FDR estimates of the software tools. One of the ways to do that is use two-species spectral libraries, wherein peptides and proteins from one of the species act as 'decoys' (but the processing software does not know that) and are not expected to be found in the sample. Two important considerations here. First, need to make sure that the peptides from the decoy species are properly annotated as not belonging to the target species, and make sure that they absolutely cannot be present in the sample and that they are not among the common contaminants. Because of this it is not recommended, for example, to use a human library as a decoy when analysing a non-human sample. Second, it is important to apply the pi0 correction when estimating false discovery rates, that is estimate the prior probability of incorrect identification, see an example here. Of note, when estimating precursor-level FDR using this method, it is absolutely essential not to apply any kind of protein q-value filtering.
Finally, an important consideration is to make sure to compare apples to apples. For example, it only makes sense to compare global protein q-value filtering in one software with that in another, and not to compare global protein q-value filtering to run-specific one.
Importantly, there is no 'one true way' to benchmark FDR, and therefore it is OK and expected that FDR estimates using controlled benchmarks can deviate from the internal FDR estimates of the software.
Q: How to compare the quantification performance of two DIA software tools?
A: The basic level at which this is often done is comparing the CV values. Or, better, comparing the numbers of precursors quantified with CVs less than a given threshold, e.g. 0.1 or 0.2. The difficulty here is that there are algorithms which can significantly improve CV values, but at the same time make quantification less reliable, because what is being integrated is not only the signal from the target precursor, but also signal from some (unknown) interfering precursor. One example is provided by DIA-NN's high accuracy and high precision modes: the high accuracy mode demonstrates better accuracy, while high precision mode yields lower CV values.
LFQbench-type experiments, featuring two mixtures of different species digests, allow to assess the accuracy of quantification. So what makes sense is to require a quantification method to yield both reasonably good accuracy and reasonably good CV values. However there is currently no known way to determine which quantification method is the best, provided the methods considered are OK in terms of both accuracy and CVs. Of note, using the number of proteins detected as differentially abundant in LFQbench as a readout does not work well: most interfering signals come from the species injected in 1:1 ratio, and therefore "integrating interferences" in this case has little to no negative effect on the ability to detect differential abundance.
Q: What happens if I process data from some related strains of my favourite organism, using a canonical spectral library?
A: If you don't have the genomes of those related strains sequenced, what might happen is that a canonical peptide in the library will get matched to a spectrum originating from a mutated peptide. For example, it can happen that PEPTIDE is matched to a spectrum originating from PNPTIDE. Will this happen often? Probably not. However if you wish to void this, Proteoforms scoring in DIA-NN is the solution.
Q: What if I don't know what kind of modifications might be there on my peptides and I want to make sure that the peptidoform is correctly identified?
A: Proteoforms scoring in DIA-NN solves this.
Q: Can I install multiple versions of DIA-NN on the same PC?
A: Yes! And this is one of the advantages of DIA-NN. You can keep the old versions for however long you need them, to enable reproducible reprocessing of old experiments, while also having the latest cutting-edge DIA-NN installed.
Q: Are DIA-NN version numbers indicative of the performance?
A: No, they are just chronological.
Q: I have a cool idea, can DIA-NN do [some magic]?
A: Maybe DIA-NN already supports it (there are some major capabilities that are implemented in DIA-NN but not yet reflected in this guide), maybe it is realistic to implement. Please email!
Please post any questions, feedback, comments or suggestions on the GitHub Discussion board (alternatively can create a GitHub issue) or email Vadim Demichev (email: my first name dot my last name at gmail dot com). General discussions on proteomics & mass specs are also welcome on the GitHub Discussion board. Can also contact me via Twitter @DemichevLab or LinkedIn. If a query is unanswered for several days, please post a reminder (and keep posting until answered :)).
For questions of the type "I ran DIA-NN on some experiment and the results are surprising/strange/not as expected, why is that?", please attach, whenever possible, the full DIA-NN log recorded at Log level = 3 (can be saved using the "Save log" button).
In most cases, the reasons for any unexpected performance can be identified with the following steps:
-
Check if DIA-NN has printed any errors or warnings (in the log window of the DIA-NN GUI) when it had finished analysing - if yes, read those and assume that these are the reason for the unexpected behaviour you observe, try to fix them (DIA-NN will often suggest how) and see if it works.
-
Check if what DIA-NN has printed immediately after being launched makes sense, i.e. could it be a typo or some other mistake which led to DIA-NN being configured incorrectly?
-
If you have changed the default settings, was this done following the recommendations in this guide? If not, try to only change settings when recommended. For example, if you've searched M(ox) as a variable modification, then, unless you are specifically interested in biological signficance of methionine oxidations introduced in vivo, try analysing without M(ox). If the max variable modifications settings was set to some high number, try reducing it to 1.
-
If you see zero or too few identifications from a particular run with a predicted library that you have generated from a sequence database, see what results you can get with an empirical spectral library instead. Can use any suitable public library or a DIA-based library generated by DIA-NN from any other runs (could be from a different platform, e.g. some public data). If still not working, try to see if it's a mass calibration problem, by searching with a wide mass window first using --mass-acc-cal 100. Further, make sure that the predicted/empirical library used reflects the background proteome of your samples, not just specific proteins of interest.
-
If DIA-NN has exited unexpectedly, could it be that it ran out of memory? Memory usage is expected to be high when (i) the search space is large, e.g. for phospho or metaproteomics searches, or when allowing lots of variable modifications - see the number of library precursors reported by DIA-NN: RAM usage to store the spectral library in memory is approximately 1Gb per 1 million of precursors. Try following the steps for reducing RAM usage outlined in Frequently asked questions (FAQ).
Please cite:
DIA-NN: neural networks and interference correction
enable deep proteome coverage in high throughput Nature Methods, 2020
Using DIA-NN for the analysis of post-translation modifications (PTMs), such as phosphorylation or ubiquitination: Time-resolved in vivo ubiquitinome profiling by DIA-MS reveals USP7 targets on a proteome-wide scale Nature Communications, 2021
Using DIA-NN's ion mobility module for timsTOF data analysis or using DIA-NN in combination with FragPipe-generated spectral libraries: dia-PASEF data analysis using FragPipe and DIA-NN for deep proteomics of low sample amounts Nature Communications, 2022
Using DIA-NN for the analysis of multiplexed samples (SILAC, mTRAQ, etc): Increasing the throughput of sensitive proteomics by plexDIA Nature Biotechnology, 2022
Using DIA-NN as part of the CysQuant workflow: CysQuant: Simultaneous quantification of cysteine oxidation and protein abundance using data dependent or independent acquisition mass spectrometry Redox Biology, 2023
Using DIA-NN's QuantUMS module for quantification: QuantUMS: uncertainty minimisation enables confident quantification in proteomics biorxiv
Using DIA-NN to process Slice-PASEF data: Slice-PASEF: fragmenting all ions for maximum sensitivity in proteomics biorxiv
Using DIA-NN as part of the MSFragger-DIA workflow in FragPipe: Analysis of DIA proteomics data using MSFragger-DIA and FragPipe computational platform Nature Communications, 2023, or, in case of timsTOF data analysis with MSFragger-DIA, diaTracer enables spectrum-centric analysis of diaPASEF proteomics data biorxiv
Using DIA-NN as part of the MSBooster workflow in FragPipe: MSBooster: improving peptide identification rates using deep learning-based features Nature Communications, 2023
Other key papers
- Using DIA-NN for large-scale plasma & serum proteomics:
Cell Systems, 2020 and Cell Systems, 2021 - Ultra-fast proteomics with DIA-NN and Scanning SWATH:
Nature Biotechnology, 2021
Visualisation of peptide positions in the protein: https://github.com/MannLabs/alphamap (AlphaMap by Mann lab)
Notes and discussions on proteomics in general and the use of DIA-NN: https://github.com/vdemichev/DiaNN/discussions/categories/dia-proteomics-in-detail (this section will be further expanded).