-
Notifications
You must be signed in to change notification settings - Fork 5
Greenness
Information about determining and interpreting the 'greenness' of PhenoCam images. More information to come soon.
The Green Chromatic Coordinate is a widely used measure of greenness in PhenoCam images. The GCC is a non-linear transformation of the camera's measured green digital numbers to values representing the proportion of the greenness measured. GCC is thus an easy to understand (think percent green) measure which directly corresponds to the changing levels of green pigmentation in vegetation. The use of GCC has also been shown to reduce the influence of differences in scene illumination between images. Indeed, Sonnentag et al. found that weather-induced changes in scene illumination were 'largely suppressed' when using GCC.
GCC is easily calculated:
GCC = Green / (Green + Red + Blue)
Note that the red and blue chromatic coordinates can likewise be calculated:
RCC = Red / (Green + Red + Blue)
BCC = Blue / (Green + Red + Blue)
The Red Chromatic Coordinate has been investigated for its use in studying autumn phenology in some environments. Sonnentag et al., for example, found that the RCC followed a pattern opposite to GCC in the autumn for Red Oak leaves.
Analysis of PhenoCam images using GCC typically involves finding the mean GCC value for a region of interest within the image. The mean GCC for a PhenoCam image can be calculated in Python as follows:
from scipy import misc
# Read a PhenoCam image
img = misc.imread('/path/to/phenocam/image.jpg')
# Extract the mean red, green, and blue values from the image bands:
red = img[:,:,0].mean()
green = img[:,:,1].mean()
blue = img[:,:,2].mean()
# Calculate GCC:
img_gcc = green / (green + red + blue)Or, using the phenocam_toolkit's get_dn_means() function:
from PIL import Image
from phenocam_toolkit import get_dn_means
# Open an RGB and roi image
img = Image.open('/path/to/RGB/PhenoCam/image.jpg')
roi = Image.open('/path/to/roi/image.tif')
# Obtain the mean red, green, and blue digital number values of the image within the ROI
[r_mean_roi, g_mean_roi, b_mean_roi] = get_dn_means(img,roi)
# Calculate the mean GCC value
masked_gcc = g_mean_roi / (r_mean_roi + g_mean_roi, + b_mean_roi)Alternatively, one can use the mean_gcc() function included in greenness.py:
from greenness import mean_gcc # greenness.py from PhenoAnalysis/Python/greenness.py
# Calculate mean GCC
img_gcc = mean_gcc(img, roi)Although GCC is a relatively strong indicator of greenness under varying illumination conditions, relying on 'raw' GCC values alone will result in GCC time series marred by noise from varied weather conditions. A common method for minimizing variance in GCC includes calculating the mean GCC over an ROI for only mid-day images. Such images tend to have less variability, day to day, than other times of the day.
This method is also broadly insufficient, however. Sonnentag et al., therefore, proposes the use of a 90th percentile method, per90. According to Sonnentag et al., taking the 90th Percentile of all daytime GCC values over a 3 day window, and assigning that value to the center day (such that there is one observation per 3 days), results in a significantly better signal that produces a 34% lower RMSE between observed GCC and fitted Loess curves.
An example of how to obtain per90 GCC using the per90() function of greenness.py is shown below. A more detailed example will come later...
from greenness import per90
'''
Given the variable 'gcc' is a list of gcc values and the
variable 'dates' is a list of datetime objects corresponding
to the date/time of the gcc values in 'gcc'
'''
per90_gcc, per90_dates = per90(dates, gcc)A figure showing raw and per90 GCC values between the hours of 10:00 AM and 2:00 PM for the kansas PhenoCam site is shown below:
About Excess greenness and its calculation. Coming soon.
The Normalized Difference Vegetation Index (NDVI) is a common measure of vegetation health. The NDVI is calculated using the red and near-infrared parts of the spectrum:
NDVI = (NIR - Red) / (NIR + Red)
For PhenoCams that include an 'IR View,' it is possible to create an NDVI image using the RGB and IR images with the same timestamps.
It should be noted that the 'IR View' available through standard Stardot SC PhenoCam cameras is obtained through the use of an IR filter. During regular RGB mode, the IR filter is in place, blocking IR radiation from striking the CCD array. While in IR mode, the filter is removed, allowing infrared radiation to reach the CCD array. The IR image is thus not a perfect comparison to other remote sensing instruments with dedicated IR sensors.
The NIR component can nevertheless be isolated from the visible component. As described by Petach et al., 2014:
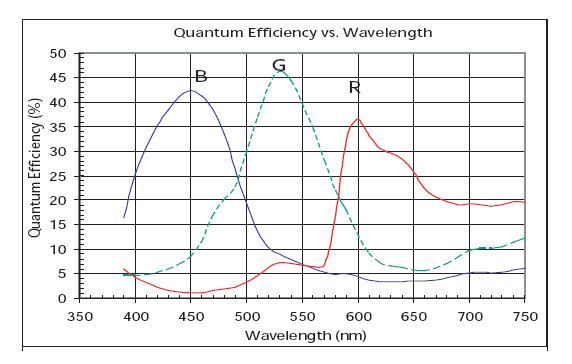
IR filter properties:
NOTE: THIS EXAMPLE DOES NOT UTILIZE THE METHOD DESCRIBED ABOVE BY Petach et al. The following calculation of NDVI can be considered a rough approximation.
Example:
Given the two PhenoCam images from the ninemileprairie site, create an NDVI image using Python.
from PIL import Image
import numpy as np
from matplotlib import pyplot as plt # For image viewing
# Open an RGB and IR PhenoCam image
rgb_img = Image.open('/path/to/rgb/PhenoCam/image.jpg')
ir_img = Image.open('/path/to/ir/PhenoCam/image.jpg')
# Get the red band from the rgb image, and open it as a numpy matrix
r,_,_ = rgb_img.split()
r = np.asarray(r, datatype=float)
# Get one of the IR image bands (all bands should be same)
ir,_,_ = ir_img.split()
ir = np.asarray(ir, datatype=float)
# Create a numpy matrix of zeros to hold the calculated NDVI values for each pixel
ndvi = np.zeros(r.size) # The NDVI image will be the same size as the input image
# Calculate NDVI
ndvi = np.true_divide(np.subtract(ir, r), np.add(ir, r))
# Display the results
plt.plot(ndvi)
plt.colorbar()
plt.show() # Results shown belowAlternatively, the create_ndvi function can be utilized to create ndvi images from PhenoCam RGB/IR pairs.
from scipy import misc
from greenness import create_ndvi
# Open the RGB and IR images
rgb = misc.imread('/path/to/rgb/PhenoCam/image.jpg')
ir = misc.imread('/path/to/IR/PhenoCam/image.jpg')
# Get the NDVI image
ndvi = create_ndvi(rgb, ir, saveto = '/path/to/save/ndvi/image.jpg')Other measures of greenness may include the Simple Vegetation Index, EVI, VARIgreen, etc.
Visually interpreting the greenness signal can provide valuable insight into the dynamics of vegetation within a PhenoCam's field of view. Nevertheless, a visual assessment provides little quantitative information with which to compare to greenness signals between seasons, or from other PhenoCam sites. Examples of important quantitative phenological information include the dates of spring onset, maximum leaf development, autumn onset, and the length of greenup and greendown.
There are a number of methods to derive such phenological transition dates from the greenness signal calculated from time series of PhenoCam images, ranging from the simple to complex. Indeed, perhaps the simplest approach is to use a thresholding method. Keenan et al. 2014 used a threshold of 33% of the annual amplitude of the greenness signal to find spring and fall transition dates. Although defining specific threshold values is dependent on camera setup and site characteristics (a different threshold value may be more appropriate for another site), their threshold derived transition dates were shown to be more highly correlated to ground-based observations of phenology than one of the most widely used methods - the sigmoid based approach.
The sigmoid approach models the green-up (spring) or green-down (fall) of the greenness signal using a sigmoidal function whose parameters are typically estimated using a non-linear least squares fit to the data (e.g., Hufkens et al., 2012). The sigmoid has been widely used to model various biological growth phenomena (see the entry on the Generalized Logistic Function in the catalog of Curves for Curve Fitting), and has been readily adopted by those examining PhenoCam greenness signals. Indeed, phenological transition dates can be derived from the fitted sigmoidal model using the same methods used to produce the MODIS phenology product, as per Zhang et al. 2003.
More complex sigmoid-based models have also been developed and used to characterize the PhenoCam greenness signal. Richardson et al., 2009, for example, utilized a method wherein two sigmoid functions were multiplied together, allowing a full season's worth of data to be modeled, rather than modeling the spring and fall separately with a single sigmoid. Other variations of the sigmoid and double sigmoid model have also been used, although most stem from the simple sigmoid model. Klosterman et al., 2014, for example, tests the use of three different sigmoid-based methods.
It must be noted that, with any of these approaches, there are significant issues that must be considered when working with greenness signals from PhenoCams. The foremost issue is that the dates derived from these models do not tend to accurately correspond to ground-based observations of phenology. Indeed, Keenan et al., 2014 investigated the relationship between leaf and canopy level vegetation traits and the greenness signal obtained from a PhenoCam. His findings show that PhenoCam based metrics of greenness tended to reach their maximum up to 2 weeks prior to maximum leaf area index. In his case, it was found that the greenness signal is a function of camera placement, leaf color, and canopy structure. The greenness signal quickly climbs to its maximum before full leaf development in part because the camera's oblique view means that the camera is observing many overlapping layers of leaves, saturating the greenness signal before full LAI is reached. In fact, an ice storm at the Harvard Forest site, which lead to a 22% decrease in summer LAI for 2009 demonstrates this fact. Although it took nearly 4 years for summer LAI to fully recover, the greenness signal for summertime remained unchanged in comparison to pre-ice storm years (Keenan et al., 2014).
This discussion highlights a very important fact: what the greenness signal truly corresponds to for any given site is dependent on camera setup and vegetation characteristics. A 'one model fits all' approach may therefore be at a serious disadvantage to models developed for the specific conditions exhibited at any given site. A careful examination of the relationship between the greenness signal derived from PhenoCams and the vegetation they monitor at sites other than Northeastern deciduous forests (where much of this research has taken place, e.g., Harvard Forest), is needed.