-
Notifications
You must be signed in to change notification settings - Fork 248
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge pull request #21 from shsarv/for-documentation
some proper documentation organisation
- Loading branch information
Showing
19 changed files
with
887 additions
and
647 deletions.
There are no files selected for viewing
This file was deleted.
Oops, something went wrong.
Binary file not shown.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,15 +1,126 @@ | ||
| # how to run this app. | ||
| - clone the repo | ||
| - move to folder BRAIN TUMOR DETECTION, | ||
| - create a virtual env using `virtualenv 'envirnoment name'.` | ||
| - Now type `. \Scripts\activate` to activate your virtualenv venv | ||
| - install packages required by the app by running the command `pip install -r requirments.txt` if you found error of torch or torch-vision libraries so your can download it from below commands. | ||
| <b><i>for windows cpu torch library command with conda</i></b>: conda install pytorch torchvision cpuonly -c pytorch | ||
| <b><i>for windows cpu torch library command with pip</i></b>: pip install torch==1.6.0+cpu torchvision==0.7.0+cpu -f https://download.pytorch.org/whl/torch_stable.html | ||
| # Brain Tumor Detection (End-to-End) | ||
|
|
||
| For Linux, Mac,CUDA version for windows and much more you can visit <a href = "https://pytorch.org/">Link</a> | ||
| - now run the app using `flask run`. | ||
| - if all things works fine so you can view running web application in your browser at port 5000. | ||
| ## Introduction | ||
|
|
||
| This project is a **Flask web application** for detecting brain tumors from MRI images using a deep learning model built with **PyTorch**. Users can upload MRI images through the app, and the model will classify them as either tumor or non-tumor. The goal of this project is to provide an intuitive interface for medical professionals to quickly identify potential brain tumors. | ||
|
|
||
| # Download the model file from [https://drive.google.com/file/d/1LJG_ITCWWtriLC5NPrWxIDwekWbhU_Rj/view?usp=sharing](https://drive.google.com/file/d/1LJG_ITCWWtriLC5NPrWxIDwekWbhU_Rj/view?usp=sharing) and add to model directory in order to run the project. | ||
| ### Dataset: | ||
| - The dataset contains MRI images, divided into two categories: **tumor** and **non-tumor**. | ||
| - Preprocessing techniques are applied to the dataset to ensure optimal model performance. | ||
|
|
||
| ## Project Overview | ||
|
|
||
| This end-to-end project consists of: | ||
| 1. **Data Loading**: Load MRI images for training, validation, and testing. | ||
| 2. **Data Preprocessing**: Apply normalization, resizing, and augmentation techniques. | ||
| 3. **Model Building**: Build a Convolutional Neural Network (CNN) using **PyTorch** to classify the MRI images. | ||
| 4. **Model Training**: Train the model on GPU (if available) to detect brain tumors. | ||
| 5. **Flask Web Application**: Develop a Flask app for user interaction, allowing image uploads for tumor detection. | ||
| 6. **Model Deployment**: Deploy the trained model within the Flask app. | ||
| 7. **Prediction**: Provide real-time predictions through the Flask web app. | ||
|
|
||
| ## Model Download and Directory Structure | ||
|
|
||
| ### Pretrained Model: | ||
| You can download the pretrained model from the following link: | ||
| [Brain Tumor Detection Model](https://drive.google.com/file/d/1LJG_ITCWWtriLC5NPrWxIDwekWbhU_Rj/view?usp=sharing) | ||
|
|
||
| ### Directory Structure: | ||
| ``` | ||
| Brain-Tumor-Detection/ | ||
| │ | ||
| ├── app/ | ||
| │ ├── static/ # CSS, JS, and images for the Flask web app | ||
| │ ├── templates/ # HTML templates for the Flask app | ||
| │ └── app.py # Main Flask application | ||
| │ | ||
| ├── model/ | ||
| │ └── brain_tumor_model.pth # Pretrained PyTorch model | ||
| │ | ||
| ├── data/ | ||
| │ ├── train/ # Training MRI images | ||
| │ ├── test/ # Testing MRI images | ||
| │ | ||
| ├── src/ | ||
| │ ├── dataset.py # Script to load and preprocess the dataset | ||
| │ ├── model.py # CNN model architecture using PyTorch | ||
| │ └── train.py # Script to train the model | ||
| │ | ||
| ├── README.md # Project documentation | ||
| └── requirements.txt # List of required Python packages | ||
| ``` | ||
|
|
||
| ## Setup Instructions | ||
|
|
||
| ### Step 1: Create a Virtual Environment | ||
|
|
||
| Create a virtual environment to isolate the dependencies for this project. | ||
|
|
||
| ```bash | ||
| # For Windows | ||
| python -m venv venv | ||
| venv\Scripts\activate | ||
|
|
||
| # For macOS/Linux | ||
| python3 -m venv venv | ||
| source venv/bin/activate | ||
| ``` | ||
|
|
||
| ### Step 2: Install Required Libraries | ||
|
|
||
| Install the dependencies listed in `requirements.txt`: | ||
|
|
||
| ```bash | ||
| pip install -r requirements.txt | ||
| ``` | ||
|
|
||
| ### Step 3: Download the Pretrained Model | ||
|
|
||
| Download the pretrained model from [this link](https://drive.google.com/file/d/1LJG_ITCWWtriLC5NPrWxIDwekWbhU_Rj/view?usp=sharing) and place it in the `model/` directory as `brain_tumor_model.pth`. | ||
|
|
||
| ### Step 4: Running the Flask App | ||
|
|
||
| To start the Flask web app, navigate to the `app/` directory and run the `app.py` file: | ||
|
|
||
| ```bash | ||
| cd app/ | ||
| python app.py | ||
| ``` | ||
|
|
||
| The app will be hosted at `http://127.0.0.1:5000/`. You can open the URL in your browser and upload MRI images to receive predictions. | ||
|
|
||
| ## Flask Web Application Features | ||
|
|
||
| - **Image Upload**: Users can upload MRI images through the web interface. | ||
| - **Tumor Detection**: The uploaded image is fed into the model to predict whether a tumor is present. | ||
| - **Result Display**: The result is displayed on the same page with either a "Tumor" or "Non-Tumor" label. | ||
|
|
||
| ## Model Architecture | ||
|
|
||
| The model used in this project is a **Convolutional Neural Network (CNN)** built using **PyTorch**. The architecture has been optimized for image classification tasks and consists of several layers: | ||
|
|
||
| ### Key Layers: | ||
| - **Convolutional Layers**: For feature extraction from MRI images. | ||
| - **Max Pooling Layers**: For downsampling and reducing spatial dimensions. | ||
| - **Fully Connected Layers**: For classification. | ||
| - **Softmax Activation**: To produce the output probability of each class (Tumor/Non-Tumor). | ||
|
|
||
| ## Data Preprocessing | ||
|
|
||
| To ensure the CNN model performs optimally, the following preprocessing steps are applied: | ||
| - **Grayscale Conversion**: All MRI images are converted to grayscale. | ||
| - **Resizing**: Images are resized to 64x64 pixels for uniformity. | ||
| - **Normalization**: Each pixel value is normalized to a range of [0, 1]. | ||
| - **Data Augmentation**: Techniques like random rotation, flipping, and zooming are applied to expand the dataset and prevent overfitting. | ||
|
|
||
| ## Conclusion | ||
|
|
||
| This Flask web app provides an end-to-end solution for detecting brain tumors using MRI images. With a simple user interface and a powerful backend, it can serve as a diagnostic tool for medical professionals. The project can be further enhanced by incorporating additional data, improving model accuracy, or deploying the app to a cloud platform like Heroku. | ||
|
|
||
| ## Future Enhancements | ||
|
|
||
| - **Integration with Cloud Platforms**: Deploy the app on Heroku or AWS for wider accessibility. | ||
| - **Mobile Application**: Develop a mobile app to upload MRI images and get predictions on the go. | ||
| - **Transfer Learning**: Incorporate pre-trained models like ResNet to further improve accuracy. | ||
|
|
||
| --- |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,38 +1,70 @@ | ||
| # Project-Arrhythmia | ||
|
|
||
| ## Introduction | ||
|
|
||
| This project focuses on predicting and classifying arrhythmias using various machine learning algorithms. The dataset used for this project is from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Arrhythmia), which consists of 452 examples across 16 different classes. Among these, 245 examples are labeled as "normal," while the remaining represent 12 different types of arrhythmias, including "coronary artery disease" and "right bundle branch block." | ||
|
|
||
| ### Introduction | ||
| ### Dataset Overview: | ||
| - **Number of Examples**: 452 | ||
| - **Number of Features**: 279 (including age, sex, weight, height, and various medical parameters) | ||
| - **Classes**: 16 total (12 arrhythmia types + 1 normal group) | ||
|
|
||
| The Dataset used in this project is available on the UCI machine learning Repository. | ||
| It can be found at: [https://archive.ics.uci.edu/ml/datasets/Arrhythmia](https://archive.ics.uci.edu/ml/datasets/Arrhythmia). | ||
| * It consists of 452 different examples spread over 16 classes. Of the 452 examples,245 are of "normal" people. | ||
| * We also have 12 different types of arrhythmias. Among all these types of arrhythmias, the most representative are the "coronary artery disease" and "Rjgbt boundle branch block". | ||
| * We have 279 features, which include age, sex, weight, height of patients and other related information. | ||
| * We explicitly observe that the number of features is relatively high compared to the number of examples we are available. | ||
| * Our goal was to predict if a person is suffering from arrhythmia or not, and if yes, classify it in to one of 12 available groups. | ||
| **Objective**: | ||
| The goal of this project is to predict whether a person is suffering from arrhythmia, and if so, classify the type of arrhythmia into one of the 12 available groups. | ||
|
|
||
| ## Algorithms Used | ||
|
|
||
| To address the classification task, the following machine learning algorithms were employed: | ||
|
|
||
| 1. **K-Nearest Neighbors (KNN) Classifier** | ||
| 2. **Logistic Regression** | ||
| 3. **Decision Tree Classifier** | ||
| 4. **Linear Support Vector Classifier (SVC)** | ||
| 5. **Kernelized Support Vector Classifier (SVC)** | ||
| 6. **Random Forest Classifier** | ||
| 7. **Principal Component Analysis (PCA)** (for dimensionality reduction) | ||
|
|
||
| ### Algorithms Used | ||
| 1. KNN Classifier | ||
| 2. Logestic Regression | ||
| 3. Decision Tree Classifier | ||
| 4. Linear SVC | ||
| 5. Kernelized SVC | ||
| 6. Random Forest Classifier | ||
| 7. Principal Component analysis (PCA) | ||
| ## Project Workflow | ||
|
|
||
| ### Result | ||
| ### Step 1: Data Exploration | ||
| - Analyzed the 279 features to identify patterns and correlations that could help with prediction. | ||
| - Addressed the challenge of the high number of features compared to the limited number of examples by employing PCA. | ||
|
|
||
|  | ||
|
|
||
| ### Step 2: Data Preprocessing | ||
| - Handled missing values, standardized data, and prepared it for machine learning models. | ||
| - Applied **Principal Component Analysis (PCA)** to reduce the feature space and eliminate collinearity, improving both execution time and model performance. | ||
|
|
||
| ### Step 3: Model Training and Evaluation | ||
| - Trained various machine learning algorithms on the dataset. | ||
| - Evaluated model performance using accuracy, recall, and other relevant metrics. | ||
|
|
||
| ### Step 4: Model Tuning with PCA | ||
| - PCA helped reduce the complexity of the dataset, leading to improved model accuracy and reduced overfitting. | ||
| - After applying PCA, models were retrained, and significant improvements were observed. | ||
|
|
||
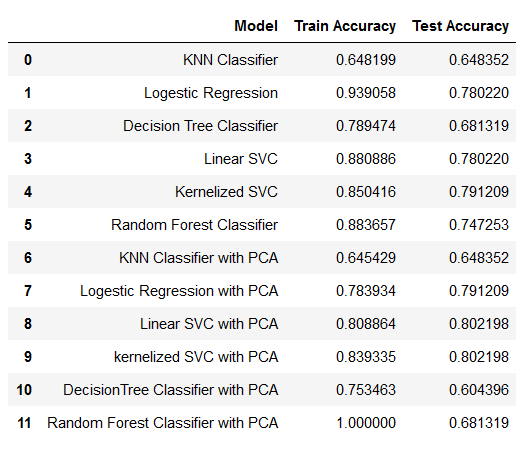
| ## Results | ||
|
|
||
|  | ||
|
|
||
| ### Conclusion | ||
|
|
||
| The models started performing better after we applied PCA on the resampled data. The reason behind this is, PCA reduces the complexity of the data. It creates components based on giving importance to variables with large variance and also the components which it creates are non collinear in nature which means it takes care of collinearity in large data set. PCA also improves the overall execution time and quality of the models and it is very beneficial when we are working with huge amount of variables. | ||
| Applying **Principal Component Analysis (PCA)** to the resampled data significantly improved the performance of the models. PCA works by creating non-collinear components that prioritize variables with high variance, thus reducing dimensionality and collinearity, which are key issues in large datasets. PCA not only enhanced the overall execution time but also improved the quality of predictions. | ||
|
|
||
| - The **best-performing model** in terms of recall score is the **Kernelized Support Vector Machine (SVM)** with PCA, achieving an accuracy of **80.21%**. | ||
|
|
||
| ## Future Work | ||
|
|
||
| - Experiment with more advanced models like **XGBoost** or **Neural Networks**. | ||
| - Perform hyperparameter tuning to further improve model accuracy and recall. | ||
| - Explore feature selection techniques alongside PCA to refine the feature set. | ||
|
|
||
|
|
||
| ## Acknowledgments | ||
|
|
||
| **The Best model in term of recall score is Kernalized SVM with PCA having accuracy of 80.21%.** | ||
| - [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Arrhythmia) | ||
| - [Scikit-learn Documentation](https://scikit-learn.org/stable/) | ||
| - [PCA Concepts](https://towardsdatascience.com/pca-using-python-scikit-learn-e653f8989e60) | ||
|
|
||
| --- | ||
|
|
||
| This `README.md` offers clear documentation of the objectives, algorithms used, results, and the significance of PCA in your project. It also provides essential information on how to run the project and the prerequisites. |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Binary file not shown.
Oops, something went wrong.