This repo contains an experiment into creating an API using RDF for integration between Sciety, bioRxiv and others.
It makes use of the Spar Ontologies, especially FRBR-aligned Bibliographic Ontology (FaBiO).
This requires Docker Compose
The Turtle files in data contain RDF statements describing articles, their versions, and their evaluations.
If you run docker-compose up --build a Trifid server is available at http://localhost:8080 to view the resources. This also has::
- a SPARQL editor at http://localhost:8080/sparql/, which uses Yasgui.
- a view on the structure at http://localhost:8080/spex/, which uses SPEX.
- a graph explorer at http://localhost:8080/graph-explorer/, which uses Graph Explorer.
This is read from a SPARQL endpoint provided by a Blazegraph instance available at http://localhost:8081/.
There is also a basic client querying and display information about articles and their evaluations at http://localhost:8082.
People think RDF is a pain because it is complicated. The truth is even worse. RDF is painfully simplistic, but it allows you to work with real-world data and problems that are horribly complicated.
Doc Maps describes 'three key requirements for representations of editorial processes in a healthy publishing ecosystem':
Extensibility: the framework should be capable of representing a wide range of editorial process events, ranging from a simple assertion that a review occurred to a complete history of editorial comments on a document to a standalone review submitted by an independent reviewer
RDF provides a simple framework for representing information; there is a wide range of existing vocabularies for describing publishing workflows, publish-able works, organisations, people, biomedical investigations...
The proposed DocMaps Framework appears to be based on JSON, which involves inventing new terms rather than being able to use existing, well-developed and tested vocabularies. Furthermore, plain JSON/XML representations usually struggle to scale to sufficiently describe the variations in the real world. (This can sometimes result in processes being limited to what the format can achieve, rather than it describing what's happening.)
Machine-readability: the framework should be represented in a format (eg XML) that can be interpreted computationally and translated into visual representations.
RDF is an abstract model that has multiple formats available. They include JSON-LD and RDF/XML, which provide access to RDF in formats already familiar to developers.
Discoverability: the framework should be publishable such that events are queryable and discoverable via a variety of well-supported mechanisms.
SPARQL is a W3C recommendation and has become the standard RDF query language.
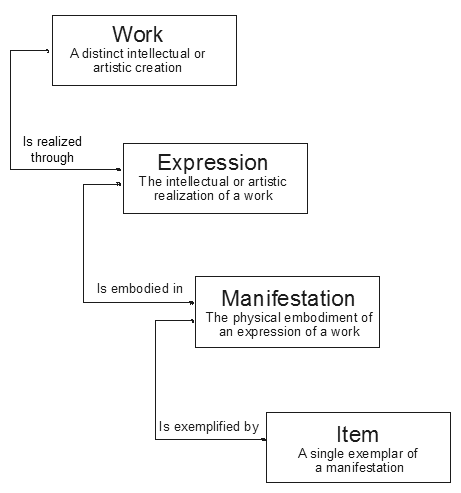
This makes use of the first 3 levels of Functional Requirements for Bibliographic Records (FRBR) in order to separate where and how an article is published from the article itself:
An article (work) can be published in multiple places (expressions), where it can have multiple formats (manifestation). Individual copies of these (item) don't need to be modelled here.
For example, for an article first published on bioRxiv then eLife:
┌───────────────────────┐
│ │
Work │ Article │
│ │
└───────────┬───────────┘
│
┌───────────────┼───────────────────────┐
│ │ │
│ │ │
┌─────▼──────┐ ┌─────▼──────┐ ┌─────▼──────┐
│ │ │ │ │ │
Expression │ bioRxiv v1 │ │ bioRxiv v2 │ │ eLife v1 │
│ │ │ │ │ │
└─────┬──────┘ └─────┬──────┘ └─────┬──────┘
│ │ │
┌────┴──┐ ┌───┴───┐ ┌───────┬───┴────┬───────┐
│ │ │ │ │ │ │ │
┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌───▼──┐ ┌──▼──┐
Manifestation │ Web │ │ PDF │ │ Web │ │ PDF │ │ Web │ │ PDF │ │ Lens │ │ ERA │
└─────┘ └─────┘ └─────┘ └─────┘ └─────┘ └─────┘ └──────┘ └─────┘
An evaluation, be it a review, recommendation or something else, links to the appropriate class.
A review by Peer Community in Ecology, for example, would apply to a single expression. That is, it covers its various formats, but not other versions of the article. A subsequent recommendation by PCI Ecology applies to both an expression and the work itself. That is, the recommendation of the article covers all other versions.
A recommendation by Peer Community in Ecology, however, could apply to the work itself. This recommendation still applies to future publication in a journal.
┌────────────────┐ ┌───────────────────────┐ ┌────────────────┐
│ PCI Ecology ├──────► ◄─────┤ eLife ◄──┐
│ Recommendation ├──┐ │ Article │ │ Recommendation │ │
└──▲─────────────┘ │ │ │ └────────┬───────┘ │
│ │ └───────────┬───────────┘ │ ┌─┴───────┐
│ └──────────┐ │ │ ┌┴────────┐│
│ ┌──────────┼────┼───────────────────────┐ │ │ eLife ││
│ │ │ │ ┌──────────────────┼──┼───────┤ Reviews ├┘
┌────────┴──┐ │ │ │ │ │ │ └─────────┘
┌┴──────────┐│ ┌─────▼──────┐ ┌▼────▼────▼─┐ ┌─────▼──▼───┐
│PCI Ecology│├───────► │ │ │ │ │
│ Reviews ├┘ │ bioRxiv v1 │ │ bioRxiv v2 │ │ eLife v1 │
└───────────┘ │ │ │ │ │ │
└─────┬──────┘ └─────┬──────┘ └─────┬──────┘
│ │ │
┌────┴──┐ ┌───┴───┐ ┌───────┬───┴────┬───────┐
│ │ │ │ │ │ │ │
┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌───▼──┐ ┌──▼──┐
│ Web │ │ PDF │ │ Web │ │ PDF │ │ Web │ │ PDF │ │ Lens │ │ ERA │
└─────┘ └─────┘ └─────┘ └─────┘ └─────┘ └─────┘ └──────┘ └─────┘
If a journal reviews submissions rather than the preprint, these are separate expressions:
┌────────────────┐ ┌───────────────────────┐ ┌────────────────┐
│ PCI Ecology ├──────► ◄─────────────┤ PeerJ ◄──┐
│ Recommendation ├──┐ │ Article │ │ Recommendation │ │
└──▲─────────────┘ │ │ │ └────────┬───────┘ │
│ │ └───────────┬───────────┘ │ ┌─┴───────┐
│ └───────────┐ │ │ ┌┴────────┐│
│ ┌───────────┼───┴────┬───────┬──────────────────┐ │ │ PeerJ ││
│ │ │ │ │ ┌────────────┼──┼───────┤ Reviews ├┘
┌────────┴──┐ │ │ │ ┌──▼─────▼────┐ │ │ └─────────┘
┌┴──────────┐│ ┌─────▼──────┐ ┌─▼────────▼─┐ ┌┴────────────┐│ ┌─────▼──▼───┐
│PCI Ecology│├───────► │ │ │ │ PeerJ ││ │ │
│ Reviews ├┘ │ bioRxiv v1 │ │ bioRxiv v2 │ │ Submissions ├┘ │ PeerJ v1 │
└───────────┘ │ │ │ │ └──────┬──────┘ │ │
└─────┬──────┘ └─────┬──────┘ │ └─────┬──────┘
│ │ ┌──▼──┐ │
┌────┴──┐ ┌───┴───┐ │ PDF │ ┌───┴───┐
│ │ │ │ └─────┘ │ │
┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐ ┌──▼──┐
│ Web │ │ PDF │ │ Web │ │ PDF │ │ Web │ │ PDF │
└─────┘ └─────┘ └─────┘ └─────┘ └─────┘ └─────┘
This graph can continue to expand with links to and from authors, reviewers, related works etc.
-
Exactly which ontologies and properties should be used/recommended? Concepts often overlap; Schema.org is well-known, but is insufficient on its own; the Spar Ontologies are great, but aren't particularly well known. Reasoning can help.
-
How should these resources be identified? Ideally by the organisation responsible (so bioRxiv for their content, eLife for their content, PCI Ecology for their content), but the work itself is only by the author(s).
-
How could this work with Doc Maps? Can be used alongside, or maybe Doc Maps should actually be RDF?