-
Notifications
You must be signed in to change notification settings - Fork 136
Illumina Nanopore read set disagreement
Unicycler produced this peculiar assembly graph for this sample:

The chromosome looks okay, and so do most plasmids. A couple of small plasmids are tangled up, but those can be resolved manually (see Tangled small plasmids). But what the heck is going on with that thing on the right?! It looks circular... could it be a plasmid too?
As in the other examples, I like to view long read alignments in Bandage to see if that sheds any light. Some reads align to a bit of the weird part of graph, but clearly originate from elsewhere in the genome:

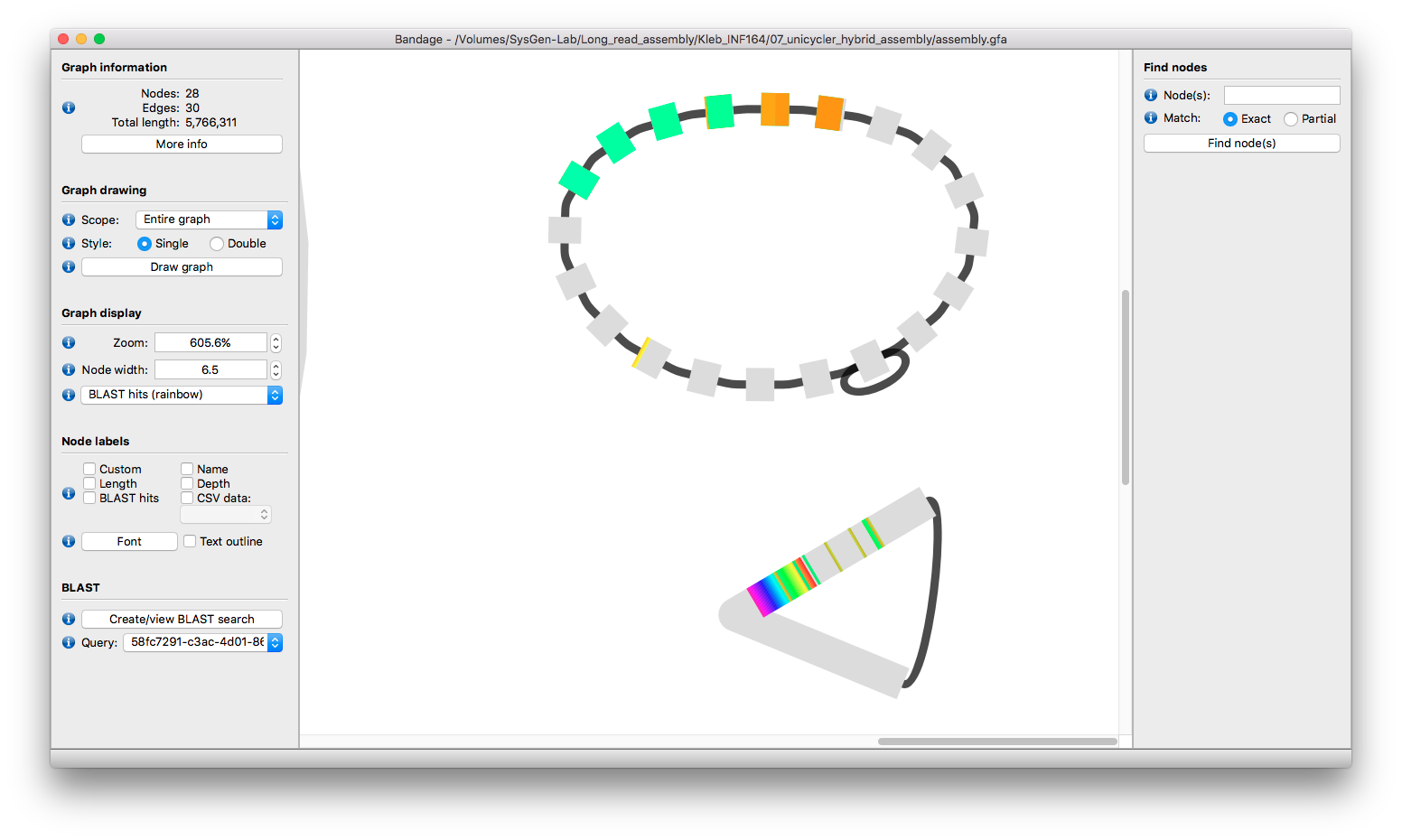
There are no reads which truly seem to 'belong' to this odd piece of the graph.
What can we draw from this? The page title may have given the answer away, but the best interpretation is that the strange part of the graph is sequence which is in the Illumina reads but not in the long reads. The genome has changed between Illumina and long read sequencing!
Unicycler works by scaffolding the Illumina graph with the long reads. Since this sequence wasn't in the long reads, it 'fell out' and was left over. Its circular appearance is not because it was a whole plasmid, but because the region has a repeat (insertion sequence) on both ends, circularising it in the graph. Unicycler doesn't handle this kind of heterogeneity well - it assumes that your read sets agree. That being said, if we delete the odd bit of the graph, we are probably left with a version of the assembly which agrees with the long reads.
The real solution is to sequence it again and assemble from consistent read sets. We did that and our second go at long read sequencing had the odd sequence included (i.e. was consistent with the Illumina reads). It turns out that it was part of the largest plasmid:
