-
Notifications
You must be signed in to change notification settings - Fork 100
Graph paths
In Bandage, a path is a means of specifying a sequence which extends through multiple nodes. You can use paths to extract sequences, and Bandage also uses paths to describe the location of BLAST queries (see BLAST searches).

Examples:
-
9+, 12-- The entirety of node 9+, followed by the entirety of node 12-
-
(51) 9+, 12-- From position 51 to the end of node 9+, followed by the entirety of node 12-
-
(51) 9+, 12- (87)- From position 51 to the end of node 9+, followed by the first 87 bases of node 12-
-
9+, 12-, 8+, 12-, 3-- This path contains a loop and includes the sequence for node 12- twice.
In Bandage, you can easily output path sequences for selected nodes.

If you wish to export the sequence for a more complex path (containing loops, start/end positions, etc.), the above approach will not work. Instead, you must select 'Specify exact path for copy/save' from the 'Output' menu.
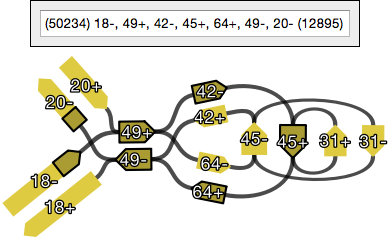
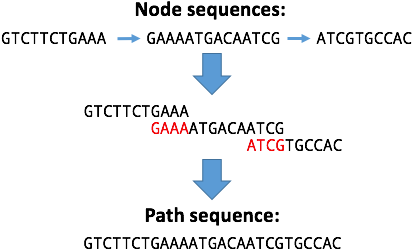
In graphs made by some assemblers, nodes connected by an edge have overlapping sequences (see assembler differences). If present, Bandage will remove this overlap when creating a path sequence. Therefore, a path sequence may be shorter than the sequences of its constituent nodes.
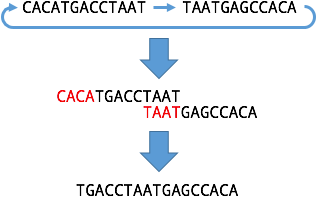
In the 'Specify exact path' window, there is a tick box for 'Circular path'. A circular path forms a loop where the sequence at the end directly leads into the sequence at the beginning. This is useful for extracting circular sequences from an assembly graph, such as bacterial chromosomes or plasmids. Circular paths, by definition, include the entirety of their constituent nodes and therefore cannot have start/end positions.

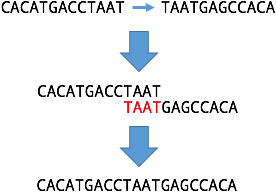
Consider two nodes which make a loop in the graph and therefore have overlaps on both ends. If you make a linear path from the two nodes, the overlap will be removed in the middle, but the start will still overlap with the end:
- Home
- Getting started
- Settings:
- Functionality:
- Assembly:
- Example uses:
- Media:
- About