This is a minimalistic, self-contained sparse Cholesky solver, supporting solving both on the CPU and on the GPU, easily integrable in your tensor pipeline.
When we were working on our "Large Steps in Inverse Rendering of Geometry" paper
[1], we found it quite challenging to hook up an existing sparse
linear solver to our pipeline, and we managed to do so by adding dependencies on
large projects (i.e. cusparse and scikit-sparse), only to use a small part
of its functionality. Therefore, we decided to implement our own library, that
serves one purpose: efficiently solving sparse linear systems on the GPU or CPU,
using a Cholesky factorization.
Under the hood, it relies on CHOLMOD for sparse matrix factorization. For the solving phase, it uses CHOLMOD for the CPU version, and uses the result of an analysis step run once when building the solver for fast solving on the GPU [2].
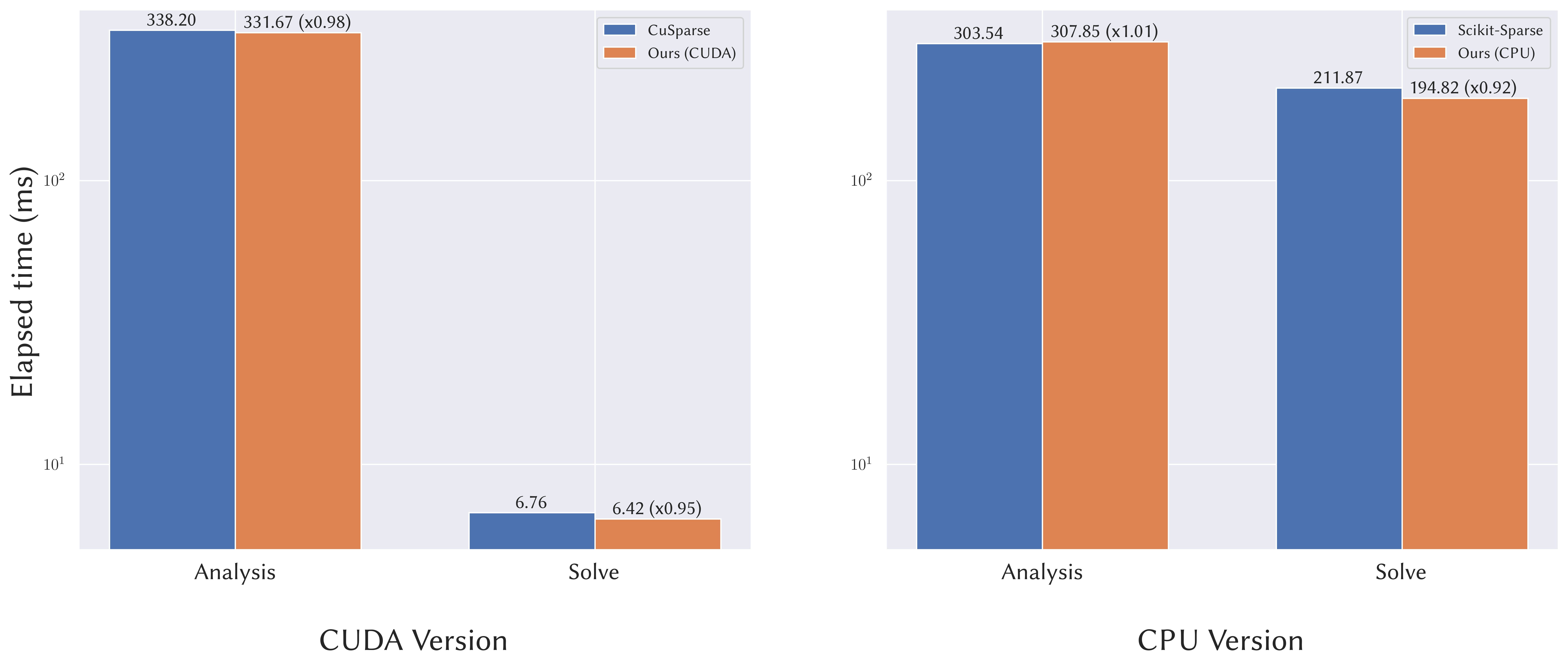
It achieves comparable performance as other frameworks, with the dependencies nicely shipped along.
Benchmark run on a Linux Ryzen 3990X workstation with a TITAN RTX.
The Python bindings are generated with nanobind, which makes it easily interoperable with most tensor frameworks (Numpy, PyTorch, JAX...)
pip install cholespygit clone --recursive https://github.com/rgl-epfl/cholespy
pip install ./cholespyThere is only one class in the module, with two variants: CholeskySolverF, CholeskySolverD. The only difference is that CholeskySolverF solves the
system in single precision while CholeskySolverD uses double precision. This
is mostly useful for solving on the GPU, as the CPU version relies on CHOLMOD,
which only supports double precision anyway.
The most common tensor frameworks (PyTorch, NumPy, TensorFlow...) are supported out of the box. You can pass them directly to the module without any need for manual conversion.
Since both variants have the same signature, we only detail CholeskySolverF
below:
cholespy.CholeskySolverF(n_rows, ii, jj, x, type)
Parameters:
n_rows- The number of rows in the (sparse) matrix.ii- The first array of indices in the sparse matrix representation. IftypeisCOO, then this is the array of row indices. If it isCSC(resp.CSR), then it is the array of column (resp. row) indices, such that row (resp. column) indices for column (resp. row)kare stored injj[ii[k]:ii[k+1]]and the corresponding entries are inx[ii[k]:ii[k+1]].jj- The second array of indices in the sparse matrix representation. IftypeisCOO, then this is the array of column indices. If it isCSC(resp.CSR), then it is the array of row (resp. column) indices.x- The array of nonzero entries.type- The matrix representation type, of typeMatrixType. Available types areMatrixType.COO,MatrixType.CSCandMatrixType.CSR.
cholespy.CholeskySolverF.solve(b, x)
Parameters
b- Right-hand side of the equation to solve. Can be a vector or a matrix. If it is a matrix, it must be of shape(n_rows, n_rhs). It must be on the same device as the tensors passed to the solver constructor. If using CUDA arrays, then the maximum supported value forn_rhsis128.x- Placeholder for the solution. It must be on the same device and have the same shape asb.
x and b must have the same dtype as the solver used, i.e. float32 for
CholeskySolverF or float64 for CholeskySolverD. Since x is modified in
place, implicit type conversion is not supported.
from cholespy import CholeskySolverF, MatrixType
import torch
# Identity matrix
n_rows = 20
rows = torch.arange(n_rows, device='cuda')
cols = torch.arange(n_rows, device='cuda')
data = torch.ones(n_rows, device='cuda')
solver = CholeskySolverF(n_rows, rows, cols, data, MatrixType.COO)
b = torch.ones(n_rows, device='cuda')
x = torch.zeros_like(b)
solver.solve(b, x)
# b = [1, ..., 1][1] Nicolet, B., Jacobson, A., & Jakob, W. (2021). Large steps in inverse rendering of geometry. ACM Transactions on Graphics (TOG), 40(6), 1-13.
[2] Naumov, M. (2011). Parallel solution of sparse triangular linear systems in the preconditioned iterative methods on the GPU. NVIDIA Corp., Westford, MA, USA, Tech. Rep. NVR-2011, 1.