 This repo contains the codebase for the attention-based implementations of VAE models for molecular design as described in this paper (note - link paper). The addition of attention allows models to learn longer range dependencies between input features and improves the quality and interpretability of learned molecular embeddings. The code is organized by folders that correspond to the following sections:
This repo contains the codebase for the attention-based implementations of VAE models for molecular design as described in this paper (note - link paper). The addition of attention allows models to learn longer range dependencies between input features and improves the quality and interpretability of learned molecular embeddings. The code is organized by folders that correspond to the following sections:
- transvae: code required to run models including model class definitions, data preparation, optimizers, etc.
- scripts: scripts for training models, generating samples and performing calculations
- notebooks: jupyter notebook tutorials and example calculations
- checkpoints: pre-trained model files
- data: token vocabularies and weights for ZINC and PubChem datasets (***note - full train and test sets for both ZINC and PubChem are available for download)
The code can be installed with pip using the following command pip install transvae. RDKit and tensor2tensor are required for certain visualizations/property calculations and must also be installed (neither of these packages are necessary for training or generating molecules so if you would prefer not to install them then you can simply remove their imports from the source code).
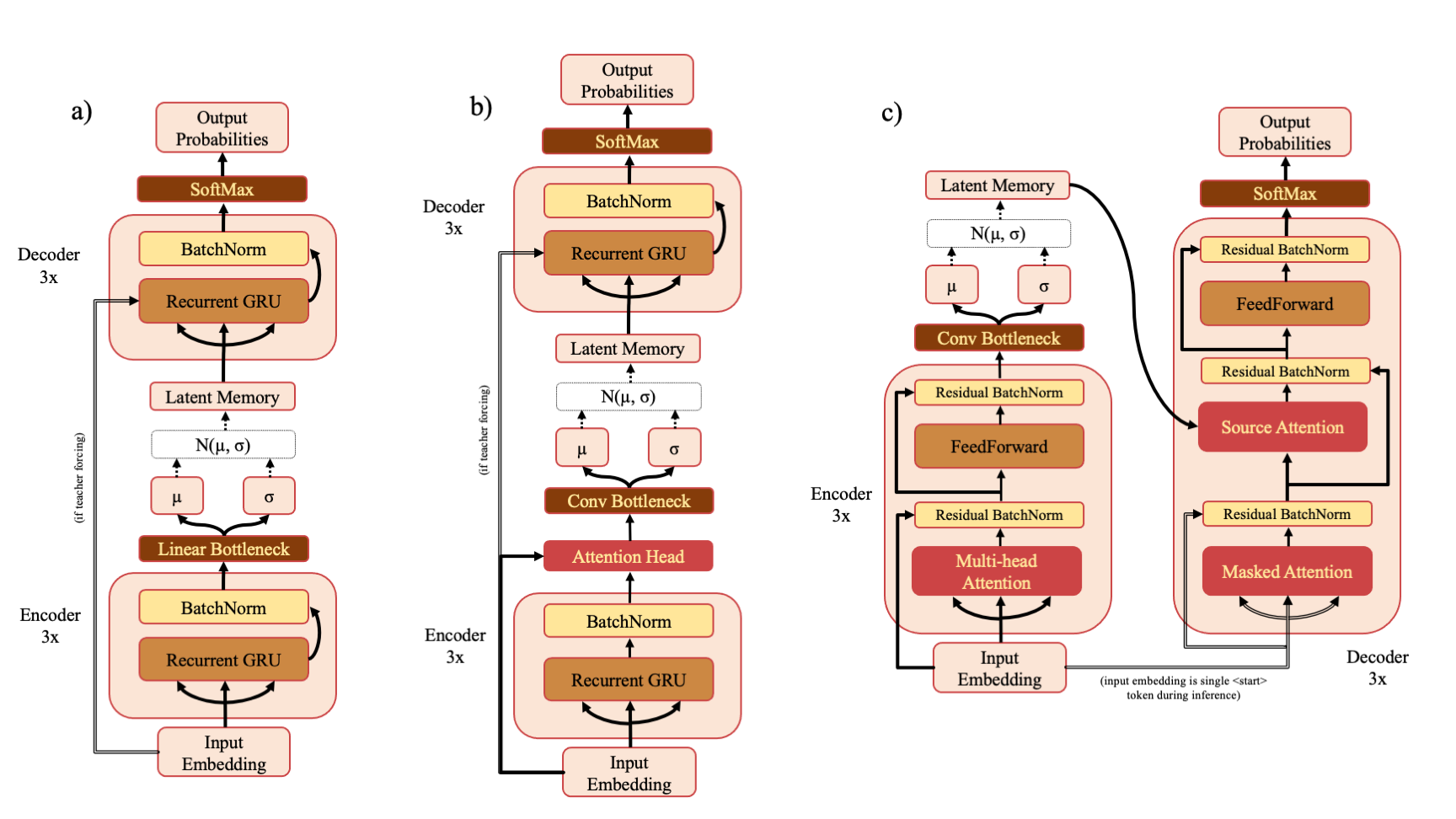
There are three model types - RNN (a), RNNAttn (b) and Transformer (c). If you've downloaded the ZINC or PubChem training sets from the drive link, you can re-train the models described in the paper with a command such as
python scripts/train.py --model transvae --data_source zinc
The default model dimension is 128 but this can also be changed at the command line
python scripts/train.py --model rnnattn --d_model 256 --data_source pubchem
You may also specify a custom train and test set like so
python scripts/train.py --model transvae --data_source custom --train_path my_train_data.txt --test_path my_test_data.txt --vocab_path my_vocab.pkl --char_weights_path my_char_weights.npy --save_name my_model
The vocabulary must be a pickle file that stores a dictionary that maps token -> token id and it must begin with the <start> or <bos> token. All modifiable hyperparameters can be viewed with python scripts/train.py --help.
There are three sampling modes to choose from - random, high entropy or k-random high entropy. If you choose to use one of the high entropy categories, you must also supply a set of SMILES (typically the training set) to use to calculate the entropy of your model prior to sampling. An example command might look like:
python scripts/sample.py --model transvae --model_ckpt checkpoints/trans4x-256_zinc.ckpt --smiles data/zinc_train.txt --sample_mode high_entropy
Attention can be calculated using the attention.py script. Due to the large number of attention heads and layers within the transvae model you should be careful about calculating attention for too many samples as it will generate a large amount of data. An example command for calculating attention might look like
python scripts/attention.py --model rnnattn --model_ckpt checkpoints/rnnattn-256_pubchem.ckpt --smiles data/pubchem_train_(n=500).txt --save_path attn_wts/rnnattn_wts.npy
Examples of model analysis functions and how to use them are shown in notebooks/visualizing_attention.ipynb and notebooks/evaluating_models.ipynb. Additionally, there are a few helper functions in transvae/analysis.py that allow you to plot training performance curves and other useful performance metrics.