- Store RDF data in Neo4j in a lossless manner (imported RDF can subsequently be exported without losing a single triple in the process).
- On-demand export property graph data from Neo4j as RDF.
- Model validation based on the W3C SHACL language
- Import of Ontologies and Taxonomies in OWL/RDFS/SKOS/...
Other features in NSMNTX include model mapping and inferencing on Neo4j graphs.
⇨ Check out the complete user manual with examples of use. ⇦
⇨ Blog on neosemantics (and more). ⇦
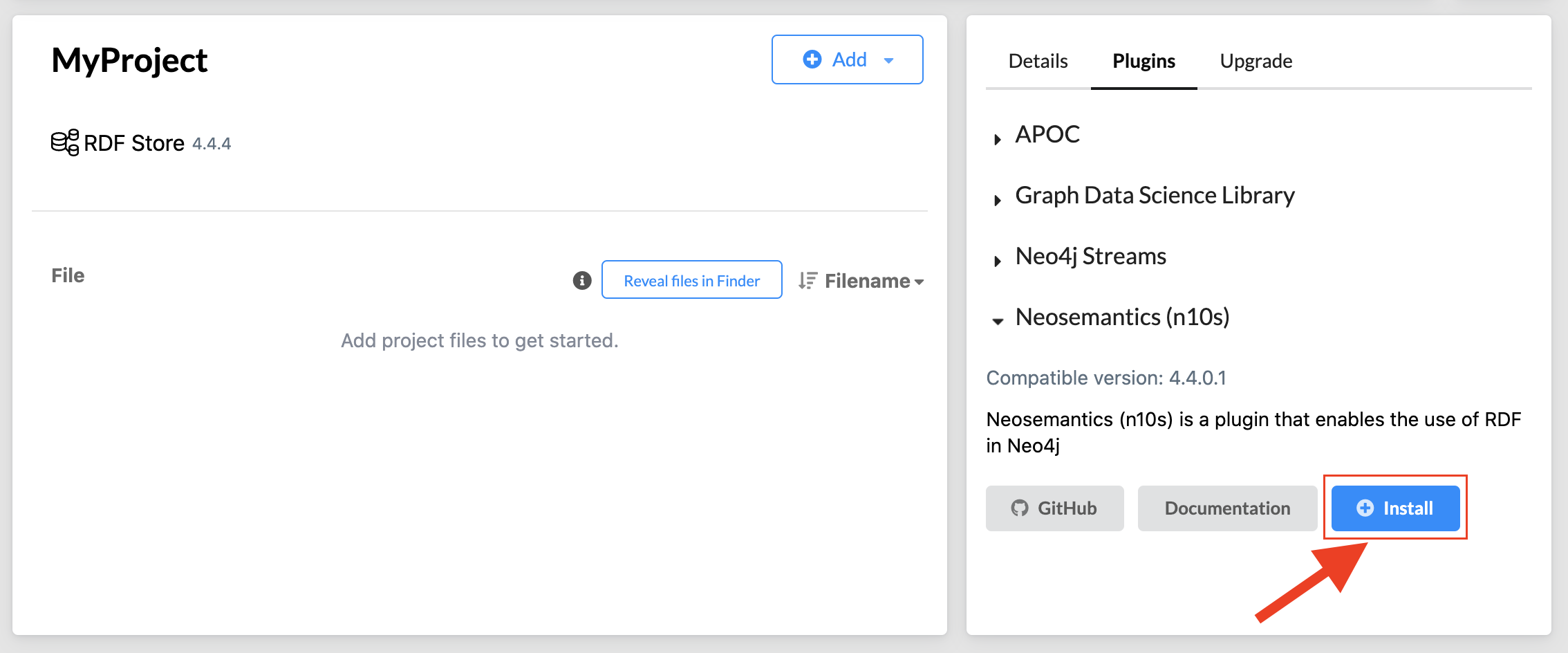
Select your database, and in the plugins section click on the install button (the most up-to-date compatible version will be shown).
If you want have the http endpoint mounted on your DB server, you'll need to add the following line to your config.
dbms.unmanaged_extension_classes=n10s.endpoint=/rdf
In the desktop you'll be able to do this by clicking on the three dots to the right hand side of your database and then select settings. You can add the fragment at the end of the file.
- Download the jar file from the releases area and copy it in the <NEO_HOME>/plugins directory of your Neo4j instance.
- Add the following line to your <NEO_HOME>/conf/neo4j.conf
dbms.unmanaged_extension_classes=n10s.endpoint=/rdf - Restart the server.
You can check that the installation went well by:
Running call dbms.procedures(). The list of procedures should include a number of them prefixed by n10s.
If you installed the http endpoint, you can check it was correctly installed by looking in the logs and making sure they show the following line on startup:
YYYY-MM-DD HH:MM:SS.000+0000 INFO Mounted unmanaged extension [n10s.endpoint] at [/rdf]
An alternative way of testing the extension is mounted is by running :GET /rdf/ping on
the neo4j browser. This should return the following message
{"ping":"here!"}
CREATE CONSTRAINT n10s_unique_uri FOR (r:Resource) REQUIRE r.uri IS UNIQUE
Before any RDF import operation a GraphConfig needs to be created. Here we define the way the RDF data is persisted in Neo4j.
We'll find things like
| Param | Values | Desc |
|---|---|---|
| handleVocabUris | "SHORTEN","KEEP","SHORTEN_STRICT","MAP" | how namespaces are handled |
| handleMultival | "OVERWRITE","ARRAY" | how multivalued properties are handled |
| handleRDFTypes | "LABELS","NODES","LABELS_AND_NODES" | how RDF datatypes are handled |
| multivalPropList | [ list of predicate uris ] | |
| ... | ... | ... |
Check the complete list in the reference.
You can create a graph config with all the defaults like this:
call n10s.graphconfig.init()
Or customize it by passing a map with your options:
call n10s.graphconfig.init( { handleMultival: "ARRAY",
multivalPropList: ["http://voc1.com#pred1", "http://voc1.com#pred2"],
keepLangTag: true })
Once the Graph config is created we can import data from a url using fetch:
call n10s.rdf.import.fetch( "https://raw.githubusercontent.com/jbarrasa/neosemantics/3.5/docs/rdf/nsmntx.ttl",
"Turtle")
Or pass it as a parameter using inline:
with '
@prefix neo4voc: <http://neo4j.org/vocab/sw#> .
@prefix neo4ind: <http://neo4j.org/ind#> .
neo4ind:nsmntx3502 neo4voc:name "NSMNTX" ;
a neo4voc:Neo4jPlugin ;
neo4voc:runsOn neo4ind:neo4j355 .
neo4ind:apoc3502 neo4voc:name "APOC" ;
a neo4voc:Neo4jPlugin ;
neo4voc:runsOn neo4ind:neo4j355 .
neo4ind:graphql3502 neo4voc:name "Neo4j-GraphQL" ;
a neo4voc:Neo4jPlugin ;
neo4voc:runsOn neo4ind:neo4j355 .
neo4ind:neo4j355 neo4voc:name "neo4j" ;
a neo4voc:GraphPlatform , neo4voc:AwesomePlatform .
' as payload
call n10s.rdf.import.inline( payload, "Turtle") yield terminationStatus, triplesLoaded, triplesParsed, namespaces
return terminationStatus, triplesLoaded, triplesParsed, namespaces
It is possible to pass some request specific parameters like headerParams, commitSize, languageFilter... (also found the reference)
Same naming scheme applies...
call n10s.onto.import.fetch(...)
Use autocompletion to discover the different procedures.
Full documentation will be available soon. In the meantime, please share your feedback in the Neo4j community portal.
Thanks!