-
Notifications
You must be signed in to change notification settings - Fork 3
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
* pruning noteboook from demo for release * - initial commit adding the predictions using pretrained model. - adding evaluation of pretrained vs course trained model - saving of the predictions - saving of the pixl based metrics and segmentation metrics * renaming the previous virtual staining demo * formatting and updaing demos readme * restructuring the folder tree * updating readme after folder reorg * - adding predictions with pretrain model - evaluation metrics pixel and segmentation - saving predictions for further evaluation * bumping cellpose to 3.0.10
- Loading branch information
1 parent
a1df436
commit baa4ee3
Showing
18 changed files
with
1,555 additions
and
68 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file was deleted.
Oops, something went wrong.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,18 @@ | ||
| # VisCy usage examples | ||
|
|
||
| Examples scripts showcasing the usage of VisCy for different computer vision tasks. | ||
|
|
||
| ## Virtual staining | ||
| ### Image-to-Image translation using VisCy | ||
| - [Guide for Virtual Staining Models](https://github.com/mehta-lab/VisCy/wiki/virtual-staining-instructions): | ||
| Instructions for how to train and run inference on ViSCy's virtual staining models (*VSCyto3D*, *VSCyto2D* and *VSNeuromast*) | ||
|
|
||
| - [Image translation Exercise](./dlmbl_exercise/solution.py): | ||
| Example showing how to use VisCy to train, predict and evaluate the VSCyto2D model. This notebook was developed for the [DL@MBL2024](https://github.com/dlmbl/DL-MBL-2024) course. | ||
|
|
||
| - [Virtual staining exercise](./img2img_translation/solution.py): exploring the label-free to fluorescence virtual staining and florescence to label-free image translation task using VisCy UneXt2. | ||
|
|
||
| ## Notes | ||
| To run the examples, make sure to activate the `viscy` environment. Follow the instructions for each demo. | ||
|
|
||
| These scripts can also be ran interactively in many IDEs as notebooks,for example in VS Code, PyCharm, and Spyder. |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
File renamed without changes.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,88 @@ | ||
| # Exercise 6: Image translation - Part 1 | ||
|
|
||
| This demo script was developed for the DL@MBL 2024 course by Eduardo Hirata-Miyasaki, Ziwen Liu and Shalin Mehta, with many inputs and bugfixes by [Morgan Schwartz](https://github.com/msschwartz21), [Caroline Malin-Mayor](https://github.com/cmalinmayor), and [Peter Park](https://github.com/peterhpark). | ||
|
|
||
|
|
||
| # Image translation (Virtual Staining) | ||
|
|
||
| Written by Eduardo Hirata-Miyasaki, Ziwen Liu, and Shalin Mehta, CZ Biohub San Francisco. | ||
|
|
||
| ## Overview | ||
|
|
||
| In this exercise, we will predict fluorescence images of nuclei and plasma membrane markers from quantitative phase images of cells, i.e., we will _virtually stain_ the nuclei and plasma membrane visible in the phase image. | ||
| This is an example of an image translation task. We will apply spatial and intensity augmentations to train robust models and evaluate their performance. Finally, we will explore the opposite process of predicting a phase image from a fluorescence membrane label. | ||
|
|
||
| [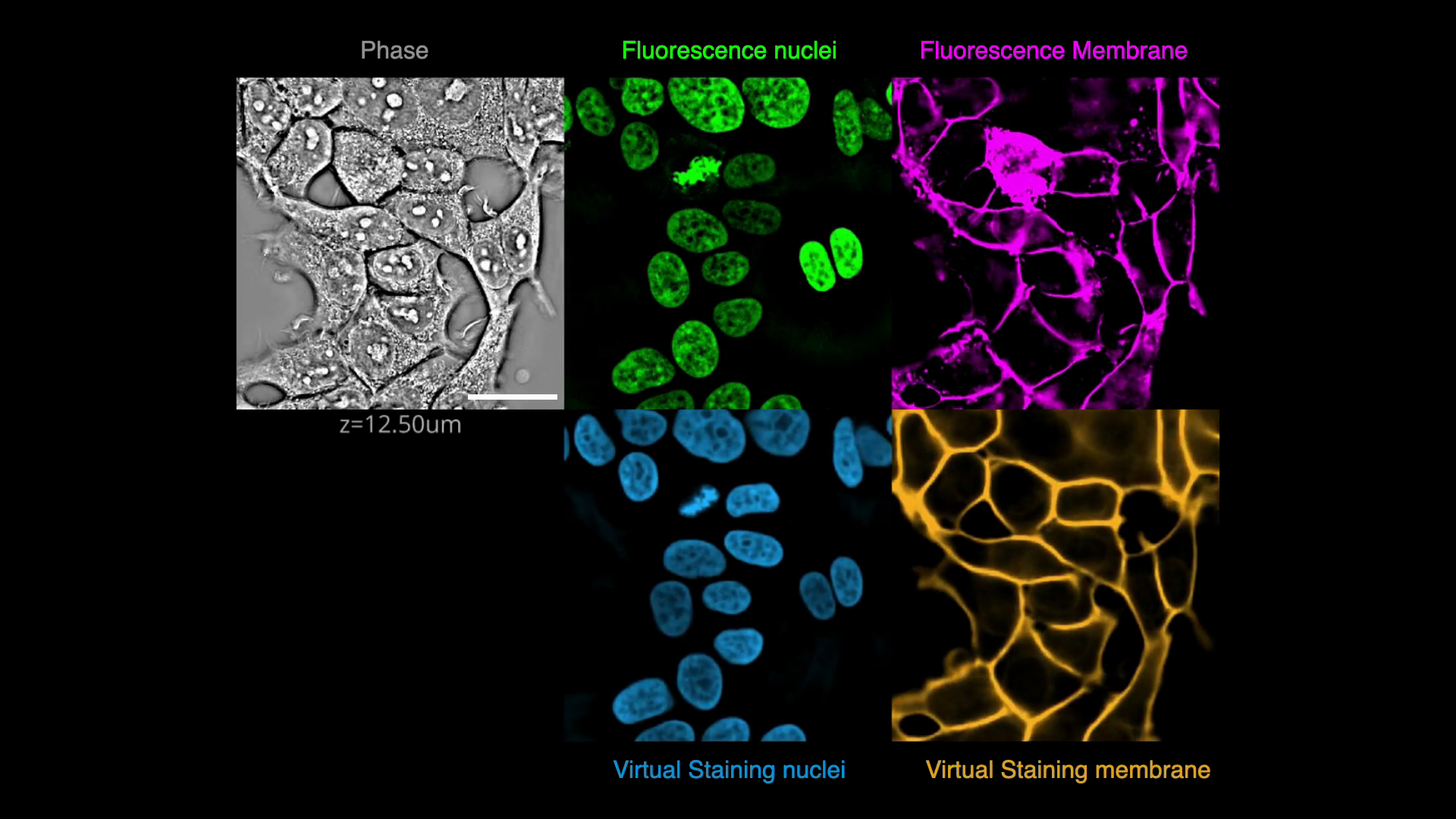](https://github.com/mehta-lab/VisCy/assets/67518483/d53a81eb-eb37-44f3-b522-8bd7bddc7755) | ||
| (Click on image to play video) | ||
|
|
||
| ## Goals | ||
|
|
||
| ### Part 1: Learn to use iohub (I/O library), VisCy dataloaders, and TensorBoard. | ||
|
|
||
| - Use a OME-Zarr dataset of 34 FOVs of adenocarcinomic human alveolar basal epithelial cells (A549), | ||
| each FOV has 3 channels (phase, nuclei, and cell membrane). | ||
| The nuclei were stained with DAPI and the cell membrane with Cellmask. | ||
| - Explore OME-Zarr using [iohub](https://czbiohub-sf.github.io/iohub/main/index.html) | ||
| and the high-content-screen (HCS) format. | ||
| - Use [MONAI](https://monai.io/) to implement data augmentations. | ||
|
|
||
| ### Part 2: Train and evaluate the model to translate phase into fluorescence, and vice versa. | ||
| - Train a 2D UNeXt2 model to predict nuclei and membrane from phase images. | ||
| - Compare the performance of the trained model and a pre-trained model. | ||
| - Evaluate the model using pixel-level and instance-level metrics. | ||
|
|
||
|
|
||
| Checkout [VisCy](https://github.com/mehta-lab/VisCy/tree/main/examples/demos), | ||
| our deep learning pipeline for training and deploying computer vision models | ||
| for image-based phenotyping including the robust virtual staining of landmark organelles. | ||
| VisCy exploits recent advances in data and metadata formats | ||
| ([OME-zarr](https://www.nature.com/articles/s41592-021-01326-w)) and DL frameworks, | ||
| [PyTorch Lightning](https://lightning.ai/) and [MONAI](https://monai.io/). | ||
|
|
||
| ## Setup | ||
|
|
||
| Make sure that you are inside of the `image_translation` folder by using the `cd` command to change directories if needed. | ||
|
|
||
| Make sure that you can use conda to switch environments. | ||
|
|
||
| ```bash | ||
| conda init | ||
| ``` | ||
|
|
||
| **Close your shell, and login again.** | ||
|
|
||
| Run the setup script to create the environment for this exercise and download the dataset. | ||
| ```bash | ||
| sh setup.sh | ||
| ``` | ||
| Activate your environment | ||
| ```bash | ||
| conda activate 06_image_translation | ||
| ``` | ||
|
|
||
| ## Use vscode | ||
|
|
||
| Install vscode, install jupyter extension inside vscode, and setup [cell mode](https://code.visualstudio.com/docs/python/jupyter-support-py). Open [solution.py](solution.py) and run the script interactively. | ||
|
|
||
| ## Use Jupyter Notebook | ||
|
|
||
| The matching exercise and solution notebooks can be found [here](https://github.com/dlmbl/image_translation/tree/28e0e515b4a8ad3f392a69c8341e105f730d204f) on the course repository. | ||
|
|
||
| Launch a jupyter environment | ||
|
|
||
| ``` | ||
| jupyter notebook | ||
| ``` | ||
|
|
||
| ...and continue with the instructions in the notebook. | ||
|
|
||
| If `06_image_translation` is not available as a kernel in jupyter, run: | ||
|
|
||
| ``` | ||
| python -m ipykernel install --user --name=06_image_translation | ||
| ``` | ||
|
|
||
| ### References | ||
|
|
||
| - [Liu, Z. and Hirata-Miyasaki, E. et al. (2024) Robust Virtual Staining of Cellular Landmarks](https://www.biorxiv.org/content/10.1101/2024.05.31.596901v2.full.pdf) | ||
| - [Guo et al. (2020) Revealing architectural order with quantitative label-free imaging and deep learning. eLife](https://elifesciences.org/articles/55502) |
3 changes: 2 additions & 1 deletion
3
examples/demo_dlmbl/convert-solution.py → ...aining/dlmbl_exercise/convert-solution.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
File renamed without changes.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,32 @@ | ||
| #!/usr/bin/env -S bash -i | ||
|
|
||
| START_DIR=$(pwd) | ||
|
|
||
| # Create conda environment | ||
| conda create -y --name 06_image_translation python=3.10 | ||
|
|
||
| # Install ipykernel in the environment. | ||
| conda install -y ipykernel nbformat nbconvert black jupytext ipywidgets --name 06_image_translation | ||
| # Specifying the environment explicitly. | ||
| # conda activate sometimes doesn't work from within shell scripts. | ||
|
|
||
| # install viscy and its dependencies`s in the environment using pip. | ||
| # Find path to the environment - conda activate doesn't work from within shell scripts. | ||
| ENV_PATH=$(conda info --envs | grep 06_image_translation | awk '{print $NF}') | ||
| $ENV_PATH/bin/pip install "viscy[metrics,visual]==0.2.0" | ||
|
|
||
| # Create the directory structure | ||
| mkdir -p ~/data/06_image_translation/training | ||
| mkdir -p ~/data/06_image_translation/test | ||
| mkdir -p ~/data/06_image_translation/pretrained_models | ||
| # Change to the target directory | ||
| cd ~/data/06_image_translation/training | ||
| # Download the OME-Zarr dataset recursively | ||
| wget -m -np -nH --cut-dirs=5 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VS_datasets/VSCyto2D/training/a549_hoechst_cellmask_train_val.zarr/" | ||
| cd ~/data/06_image_translation/test | ||
| wget -m -np -nH --cut-dirs=5 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VS_datasets/VSCyto2D/test/a549_hoechst_cellmask_test.zarr/" | ||
| cd ~/data/06_image_translation/pretrained_models | ||
| wget -m -np -nH --cut-dirs=5 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VS_models/VSCyto2D/VSCyto2D/epoch=399-step=23200.ckpt" | ||
|
|
||
| # Change back to the starting directory | ||
| cd $START_DIR |
Oops, something went wrong.