-
-
Notifications
You must be signed in to change notification settings - Fork 46
Use shinylive to show the app on the README #1490
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Open
llrs-roche
wants to merge
23
commits into
main
Choose a base branch
from
1404_shinylive_readme@main
base: main
Could not load branches
Branch not found: {{ refName }}
Loading
Could not load tags
Nothing to show
Loading
Are you sure you want to change the base?
Some commits from the old base branch may be removed from the timeline,
and old review comments may become outdated.
Open
Changes from all commits
Commits
Show all changes
23 commits
Select commit
Hold shift + click to select a range
55b587c
Transform the README.md to README.Rmd
llrs-roche f666b0d
Update readme and push it
llrs-roche 1c6064d
[skip style] [skip vbump] Restyle files
github-actions[bot] 686e5e9
Modify syntax
llrs-roche 3d61c77
Merge branch '1404_shinylive_readme@main' of https://github.com/insig…
llrs-roche 8b22630
remove renv
llrs-roche 7f9c46d
Add dependencies
llrs-roche 4b1adc7
Add hook for README
llrs-roche 810fd50
Merge branch 'main' into 1404_shinylive_readme@main
m7pr 7950690
Merge branch 'main' into 1404_shinylive_readme@main
m7pr 5c6fdb6
Remove iframe
llrs-roche 1bb91e2
Merge branch 'main' into 1404_shinylive_readme@main
llrs-roche dc0d190
Lower level of the section
llrs-roche 2355c6b
Merge upstream
llrs-roche 1b6eefc
Merged origin/main into 1404_shinylive_readme@main
llrs-roche 7dc23a7
Merge branch 'main' into 1404_shinylive_readme@main
llrs-roche 0ad3599
Update README to show the shinylive app
llrs-roche bd029e8
Consequences of the update on the README.Rmd
llrs-roche a42d4e5
[skip style] [skip vbump] Restyle files
github-actions[bot] f0fd0ec
Fix styling and add fixed id to the iframe
llrs-roche 950fec2
Merge branch '1404_shinylive_readme@main' of github.com:insightsengin…
llrs-roche cc49c62
Use ie action
llrs-roche 56ad887
Add version number to action and remove git user
llrs-roche File filter
Filter by extension
Conversations
Failed to load comments.
Loading
Jump to
Jump to file
Failed to load files.
Loading
Diff view
Diff view
There are no files selected for viewing
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -46,3 +46,4 @@ coverage.* | |
| ^.revdeprefs\.yaml$ | ||
| ^revdep$ | ||
| ^\.covrignore$ | ||
| ^README\.Rmd$ | ||
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -34,8 +34,36 @@ on: | |
| workflow_dispatch: | ||
|
|
||
| jobs: | ||
| README: | ||
| runs-on: ubuntu-latest | ||
| env: | ||
| GITHUB_PAT: ${{ secrets.GITHUB_TOKEN }} | ||
| permissions: | ||
| contents: write | ||
| steps: | ||
| - name: Checkout repo | ||
| uses: actions/[email protected] | ||
| with: | ||
| fetch-depth: 0 | ||
|
|
||
| - uses: r-lib/actions/setup-pandoc@v2 | ||
|
|
||
| - uses: r-lib/actions/setup-r@v2 | ||
|
|
||
| - uses: insightsengineering/setup-r-dependencies@v1 | ||
|
|
||
| - name: Render Rmarkdown files 🖹 | ||
| run: | | ||
| RMD_PATH=($(git diff --name-only ${{ github.event.before }} ${{ github.sha }} *.Rmd)) | ||
| Rscript -e 'for (f in commandArgs(TRUE)) if (file.exists(f)) rmarkdown::render(f)' ${RMD_PATH[*]} | ||
|
|
||
| - name: Commit results ⬆️ | ||
| run: | | ||
| git commit ${RMD_PATH[*]/.Rmd/.md} -m 'Re-build Rmarkdown files' || echo "No changes to commit" | ||
| git push origin || echo "No changes to commit" | ||
| docs: | ||
| name: Pkgdown Docs 📚 | ||
| needs: README | ||
| uses: insightsengineering/r.pkg.template/.github/workflows/pkgdown.yaml@main | ||
| secrets: | ||
| REPO_GITHUB_TOKEN: ${{ secrets.REPO_GITHUB_TOKEN }} | ||
|
|
||
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,183 @@ | ||
| --- | ||
| output: github_document | ||
| always_allow_html: true | ||
| --- | ||
|
|
||
| <!-- README.md is generated from README.Rmd. Please edit that file --> | ||
|
|
||
| ```{r, include = FALSE} | ||
| knitr::opts_chunk$set( | ||
| collapse = TRUE, | ||
| comment = "#>", | ||
| fig.path = "man/figures/README-", | ||
| out.width = "100%" | ||
| ) | ||
| ``` | ||
|
|
||
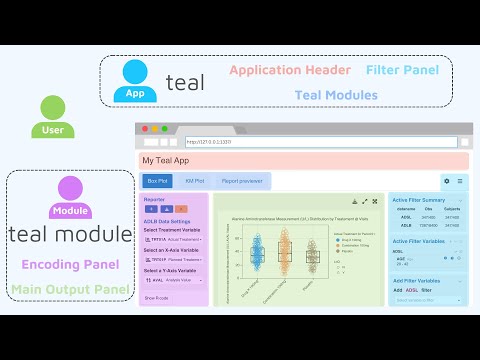
| # `teal`: Interactive Exploratory Data Analysis with `Shiny` Web-Applications <a href='https://insightsengineering.github.io/teal/'><img src="man/figures/logo.svg" align="right" height="139" style="max-width: 100%; max-height: 139px;"/></a > | ||
|
|
||
| <!-- start badges --> | ||
| [](https://cran.r-project.org/package=teal) | ||
| [](https://cran.r-project.org/package=teal) | ||
| [](https://cran.r-project.org/package=teal) | ||
| [](https://cran.r-project.org/package=teal) | ||
|
|
||
| [](https://insightsengineering.github.io/teal/main/unit-test-report/) | ||
| [](https://insightsengineering.github.io/teal/) | ||
| [](https://insightsengineering.github.io/teal/main/coverage-report/) | ||
|
|
||
|  | ||
|  | ||
|
|
||
|  | ||
|  | ||
|  | ||
|  | ||
|  | ||
|  | ||
| [](https://www.repostatus.org/#active) | ||
| [](https://github.com/insightsengineering/teal/tree/main) | ||
| [](https://github.com/insightsengineering/teal/issues?q=is%3Aissue+is%3Aopen+sort%3Aupdated-desc) | ||
| <!-- end badges --> | ||
|
|
||
| `teal` is a `shiny`-based interactive exploration framework for analyzing data. `teal` applications require app developers to specify: | ||
|
|
||
| <!-- markdownlint-disable MD007 MD030 --> | ||
| - Data, which can be: | ||
| - CDISC data, commonly used for clinical trial reporting | ||
| - Independent datasets, for example from a `data.frame` | ||
| - Related datasets, for example a set of `data.frames` with key columns to enable data joins | ||
| - `MultiAssayExperiment` objects which are `R` data structures for representing and analyzing multi-omics experiments | ||
| - `teal` modules: | ||
| - `teal modules` are `shiny` modules built within the `teal` framework that specify analysis to be performed. For example, it can be a module for exploring outliers in the data, or a module for visualizing the data in line plots. Although these can be created from scratch, many `teal` modules have been released and we recommend starting with modules found in the following packages: | ||
| - [`teal.modules.general`](https://insightsengineering.github.io/teal.modules.general/latest-tag/): general modules for exploring relational/independent/CDISC data | ||
| - [`teal.modules.clinical`](https://insightsengineering.github.io/teal.modules.clinical/latest-tag/): modules specific to CDISC data and clinical trial reporting | ||
| - [`teal.modules.hermes`](https://insightsengineering.github.io/teal.modules.hermes/latest-tag/): modules for analyzing `MultiAssayExperiment` objects | ||
|
|
||
| <!-- markdownlint-enable MD007 MD030 --> | ||
|
|
||
| A lot of the functionality of the `teal` framework derives from the following packages: | ||
|
|
||
| <!-- markdownlint-disable MD007 MD030 --> | ||
| - [`teal.data`](https://insightsengineering.github.io/teal.data/latest-tag/): creating and loading the data needed for `teal` applications. | ||
| - [`teal.widgets`](https://insightsengineering.github.io/teal.widgets/latest-tag/): `shiny` components used within `teal`. | ||
| - [`teal.slice`](https://insightsengineering.github.io/teal.slice/latest-tag/): provides a filtering panel to allow filtering of data. | ||
| - [`teal.code`](https://insightsengineering.github.io/teal.code/latest-tag/): handles reproducibility of outputs. | ||
| - [`teal.logger`](https://insightsengineering.github.io/teal.logger/latest-tag/): standardizes logging within `teal` framework. | ||
| - [`teal.reporter`](https://insightsengineering.github.io/teal.reporter/latest-tag/): allows `teal` applications to generate reports. | ||
|
|
||
| Dive deeper into `teal` with our comprehensive video guide. | ||
| Please click the image below to start learning: | ||
|
|
||
| [](https://www.youtube.com/watch?v=N8ZamECICSI) | ||
|
|
||
| <!-- markdownlint-enable MD007 MD030 --> | ||
|
|
||
| ## Installation | ||
|
|
||
| ```{r, eval=FALSE} | ||
| install.packages("teal") | ||
| ``` | ||
|
|
||
| Alternatively, you might also use the development version. | ||
|
|
||
| ```{r, eval=FALSE} | ||
| # install.packages("pak") | ||
| pak::pak("insightsengineering/teal") | ||
| ``` | ||
|
|
||
| ## Usage | ||
|
|
||
| ```{r teal, eval=FALSE} | ||
| library(teal) | ||
|
|
||
| app <- init( | ||
| data = teal_data(iris = iris), | ||
| modules = list( | ||
| module( | ||
| label = "iris histogram", | ||
| server = function(input, output, session, data) { | ||
| updateSelectInput( | ||
| session = session, | ||
| inputId = "var", | ||
| choices = names(data()[["iris"]])[1:4] | ||
| ) | ||
|
|
||
| output$hist <- renderPlot({ | ||
| req(input$var) | ||
| hist(x = data()[["iris"]][[input$var]]) | ||
| }) | ||
| }, | ||
| ui = function(id) { | ||
| ns <- NS(id) | ||
| list( | ||
| selectInput( | ||
| inputId = ns("var"), | ||
| label = "Column name", | ||
| choices = NULL | ||
| ), | ||
| plotOutput(outputId = ns("hist")) | ||
| ) | ||
| } | ||
| ) | ||
| ) | ||
| ) | ||
|
|
||
| shinyApp(app$ui, app$server) | ||
| ``` | ||
|
|
||
| ### Try it out in Shinylive | ||
|
|
||
| ```{r shinylive_url, echo = FALSE, results = 'asis', eval = requireNamespace("roxy.shinylive", quietly = TRUE)} | ||
| code <- paste0(c( | ||
| knitr::knit_code$get("teal") | ||
| ), collapse = "\n") | ||
|
|
||
| url <- roxy.shinylive::create_shinylive_url(code) | ||
| cat(sprintf("[Open in Shinylive](%s)\n\n", url)) | ||
| ``` | ||
|
|
||
| ```{r shinylive_iframe, echo = FALSE, eval = requireNamespace("roxy.shinylive", quietly = TRUE)} | ||
| # code from https://github.com/RinteRface/shinyMobile/blob/90e09384c040ba69c14293a49fc182e8d6ef26a8/R/utils.R#L232 | ||
| bslib::card( | ||
| id = "shinylive-iframe", | ||
| htmltools::tags$iframe( | ||
| class = "html-fill-item", | ||
| src = paste0(url, "&h=0"), | ||
| width = "100%", | ||
| height = 600, | ||
| allowfullscreen = "", | ||
| allow = "autoplay", | ||
| `data-external` = "1" | ||
| ), | ||
| full_screen = TRUE | ||
| ) | ||
| ``` | ||
|
|
||
| ## More resources | ||
|
|
||
| Please see [`teal.gallery`](https://insightsengineering.github.io/teal.gallery/) and [TLG Catalog](https://insightsengineering.github.io/tlg-catalog/) to see examples of `teal` apps. | ||
|
|
||
| Please start with the ["Technical Blueprint" article](https://insightsengineering.github.io/teal/latest-tag/articles/blueprint/index.html), ["Getting Started" article](https://insightsengineering.github.io/teal/latest-tag/articles/getting-started-with-teal.html), and then other [package vignettes](https://insightsengineering.github.io/teal/latest-tag/articles/index.html) for more detailed guide. | ||
|
|
||
| ## Getting help | ||
|
|
||
| If you encounter a bug or have a feature request, please file an issue. For questions, discussions, and updates, use the `teal` channel in the [`pharmaverse` slack workspace](https://pharmaverse.slack.com). | ||
|
|
||
| ## Acknowledgment | ||
|
|
||
| This package is a result of a joint efforts by many developers and stakeholders. We would like to thank everyone who contributed so far! | ||
|
|
||
| ## Stargazers and Forkers | ||
|
|
||
| ### Stargazers over time | ||
|
|
||
| [](https://starchart.cc/insightsengineering/teal) | ||
|
|
||
| ### Stargazers | ||
|
|
||
| [](https://github.com/insightsengineering/teal/stargazers) | ||
|
|
||
| ### Forkers | ||
|
|
||
| [](https://github.com/insightsengineering/teal/network/members) | ||
Oops, something went wrong.
Oops, something went wrong.
Add this suggestion to a batch that can be applied as a single commit.
This suggestion is invalid because no changes were made to the code.
Suggestions cannot be applied while the pull request is closed.
Suggestions cannot be applied while viewing a subset of changes.
Only one suggestion per line can be applied in a batch.
Add this suggestion to a batch that can be applied as a single commit.
Applying suggestions on deleted lines is not supported.
You must change the existing code in this line in order to create a valid suggestion.
Outdated suggestions cannot be applied.
This suggestion has been applied or marked resolved.
Suggestions cannot be applied from pending reviews.
Suggestions cannot be applied on multi-line comments.
Suggestions cannot be applied while the pull request is queued to merge.
Suggestion cannot be applied right now. Please check back later.
Uh oh!
There was an error while loading. Please reload this page.