Designed specifically for biological research, BioPano is a software platform targeted for visualisation of biological relationships as well as cooperative net-building. You can set up your own network of biological relationship by adding information about genetic regulation and metabolic system to the network we already provided for you. More Information please refer to our website.
View the release pages, download the corresponding platform software. Just run it!
We are sorry that we DO NOT provide a Linux version, because Adobe AIR Runtime for Linux is out of date for a long time. But you can run the .exe file with WINE.
We do not recommend users to build a local Biopano server, for one of the main advantages of Biopano is data sharing. However, for special cases such as intranet, you could use the docker repository we provide to deploy a Biopano server rapidly. We encapsulate basic databases to offer powerful basic biological data support for your intranet server. Please refer to installation for server(developing)
In Biopano, every biology part of E.coli K-12 will be displayed as a node of different types and regulation relationship will be displayed as arc of different types. They are displayed in the biological network in a visualizational way. You just need to select a node and click “Expand”, and the nodes associated with it will be “expanded”. Nodes are connected to each other by all kinds of relationship, so users can see how biological parts are connected clearly, such as LacI Operon.
However, since there are so many biological parts connected to each other in various convoluted, the expansion of all nodes will give rise to extremely involved nets, which will fail to convey useful information to the users, even interfere their thoughts. So you can choose to expand the nodes in a way you see fit. Take Transcription factor CRP as an example.
Biopano displays the network dynamically, which makes you able to tease out the relationship among thousands of materials of E.coli K-12, satisfies your curiosity and helps you discover unknown biological field.
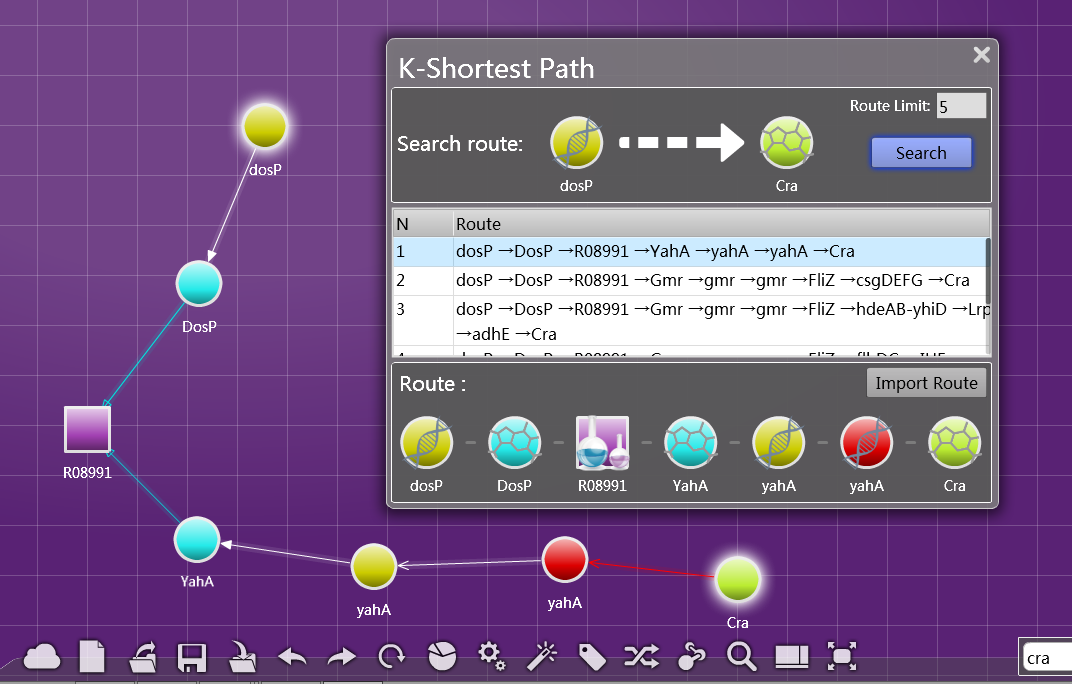
You can input two nodes that seem irrelevant, such as gene A and Transcription FactorB, and the software will search the route connecting the two nodes for you. Since some routes are too sinuous for further analysis or does not make sense, you can set a specific number k, Biopano will search and show the k shortest routes for you.
By BLAST method, Biopano finds E.coli K-12 gene highly similar to the injected sequence. These genes are regulated by other parts in E.coli K-12, so can provide information of the host environment’s impact on injected exogenous sequence. Biopano also offers BioBrick helper, enabling you search all kinds of BioBricks on iGEM’s official website. By BLAST analysis, it helps you design correct gene route with appropriate BioBricks.
Hence, while offering various functions, Biopano also serves as a cooperative network building platform. When you create a project, you can add new nodes, name new biological parts and link them with respect to your comprehension to build a brand new net gradually. Meanwhile, Biopano provides log in service. You can sign up with Google or Baidu account and upload your network to the main database to make it more plentiful. Biopano also supports data importing in batch. You can even import data of a species and a database.
When you have already got the entire view and want to know more about the details, double-click the nodes and arcs and the details will be shown. More surprisingly, Biopano supports literature reading in the network. You can view literature corresponding to every node and arc along any gene route in the network, and you will understand how the nodes in the net are associated with each other in depth. If you cast doubt on the reliability of our database, you can verify your thought with authoritative literature.
Software Usage: User Manual
bug tracker: YouTrack
Continuous Integration System: Jenkins
Code Platform: GitLab
- Yuechuan Xue [email protected]
- Shensen Zhao [email protected]
- Long Zhou [email protected]
- Yifan Gao [email protected]
- Golden Boss [email protected]
- Taixin Lin [email protected]
- Longzhi Gan [email protected]
- Yichen Fei [email protected]
- Huayu Zhang [email protected]
- Luxin Han [email protected]
- Wenshuo Wang [email protected]
- Chenxiao Dong [email protected]
- Zhiyuan Zhang [email protected]
- Tianlong Chen [email protected]
- Chuanpin Yu [email protected]
- Linfan Chen [email protected]
Biopano server is written by python, using Django Framework. The client is written by ActionScript. We use Git as our version control system. Welcome to participate in the development!
Please follow this page.
refer to the doc
Any question? Please tell us: