generated from espm-157/fish-python-template
-
Notifications
You must be signed in to change notification settings - Fork 2
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
0 parents
commit 947e1aa
Showing
7 changed files
with
430 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,21 @@ | ||
| on: push | ||
| name: Reproducibility Check | ||
|
|
||
| jobs: | ||
| render: | ||
| name: execute notebook | ||
| runs-on: arc-runner-espm157-f24 # ubuntu-latest | ||
| env: | ||
| GITHUB_PAT: ${{ secrets.GITHUB_TOKEN }} | ||
| steps: | ||
| - uses: actions/checkout@v4 | ||
| - uses: actions/setup-python@v5 | ||
| with: | ||
| python-version: '3.11.9' | ||
| - name: Install Dependencies | ||
| run: pip install -r requirements.txt | ||
| - name: test | ||
| run: | | ||
| pytest --nbval-lax *.ipynb | ||
| # - name: render | ||
| # run: myst build climate.md --pdf |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,162 @@ | ||
| # Byte-compiled / optimized / DLL files | ||
| __pycache__/ | ||
| *.py[cod] | ||
| *$py.class | ||
|
|
||
| # C extensions | ||
| *.so | ||
|
|
||
| # Distribution / packaging | ||
| .Python | ||
| build/ | ||
| develop-eggs/ | ||
| dist/ | ||
| downloads/ | ||
| eggs/ | ||
| .eggs/ | ||
| lib/ | ||
| lib64/ | ||
| parts/ | ||
| sdist/ | ||
| var/ | ||
| wheels/ | ||
| share/python-wheels/ | ||
| *.egg-info/ | ||
| .installed.cfg | ||
| *.egg | ||
| MANIFEST | ||
|
|
||
| # PyInstaller | ||
| # Usually these files are written by a python script from a template | ||
| # before PyInstaller builds the exe, so as to inject date/other infos into it. | ||
| *.manifest | ||
| *.spec | ||
|
|
||
| # Installer logs | ||
| pip-log.txt | ||
| pip-delete-this-directory.txt | ||
|
|
||
| # Unit test / coverage reports | ||
| htmlcov/ | ||
| .tox/ | ||
| .nox/ | ||
| .coverage | ||
| .coverage.* | ||
| .cache | ||
| nosetests.xml | ||
| coverage.xml | ||
| *.cover | ||
| *.py,cover | ||
| .hypothesis/ | ||
| .pytest_cache/ | ||
| cover/ | ||
|
|
||
| # Translations | ||
| *.mo | ||
| *.pot | ||
|
|
||
| # Django stuff: | ||
| *.log | ||
| local_settings.py | ||
| db.sqlite3 | ||
| db.sqlite3-journal | ||
|
|
||
| # Flask stuff: | ||
| instance/ | ||
| .webassets-cache | ||
|
|
||
| # Scrapy stuff: | ||
| .scrapy | ||
|
|
||
| # Sphinx documentation | ||
| docs/_build/ | ||
|
|
||
| # PyBuilder | ||
| .pybuilder/ | ||
| target/ | ||
|
|
||
| # Jupyter Notebook | ||
| .ipynb_checkpoints | ||
|
|
||
| # IPython | ||
| profile_default/ | ||
| ipython_config.py | ||
|
|
||
| # pyenv | ||
| # For a library or package, you might want to ignore these files since the code is | ||
| # intended to run in multiple environments; otherwise, check them in: | ||
| # .python-version | ||
|
|
||
| # pipenv | ||
| # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control. | ||
| # However, in case of collaboration, if having platform-specific dependencies or dependencies | ||
| # having no cross-platform support, pipenv may install dependencies that don't work, or not | ||
| # install all needed dependencies. | ||
| #Pipfile.lock | ||
|
|
||
| # poetry | ||
| # Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control. | ||
| # This is especially recommended for binary packages to ensure reproducibility, and is more | ||
| # commonly ignored for libraries. | ||
| # https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control | ||
| #poetry.lock | ||
|
|
||
| # pdm | ||
| # Similar to Pipfile.lock, it is generally recommended to include pdm.lock in version control. | ||
| #pdm.lock | ||
| # pdm stores project-wide configurations in .pdm.toml, but it is recommended to not include it | ||
| # in version control. | ||
| # https://pdm.fming.dev/latest/usage/project/#working-with-version-control | ||
| .pdm.toml | ||
| .pdm-python | ||
| .pdm-build/ | ||
|
|
||
| # PEP 582; used by e.g. github.com/David-OConnor/pyflow and github.com/pdm-project/pdm | ||
| __pypackages__/ | ||
|
|
||
| # Celery stuff | ||
| celerybeat-schedule | ||
| celerybeat.pid | ||
|
|
||
| # SageMath parsed files | ||
| *.sage.py | ||
|
|
||
| # Environments | ||
| .env | ||
| .venv | ||
| env/ | ||
| venv/ | ||
| ENV/ | ||
| env.bak/ | ||
| venv.bak/ | ||
|
|
||
| # Spyder project settings | ||
| .spyderproject | ||
| .spyproject | ||
|
|
||
| # Rope project settings | ||
| .ropeproject | ||
|
|
||
| # mkdocs documentation | ||
| /site | ||
|
|
||
| # mypy | ||
| .mypy_cache/ | ||
| .dmypy.json | ||
| dmypy.json | ||
|
|
||
| # Pyre type checker | ||
| .pyre/ | ||
|
|
||
| # pytype static type analyzer | ||
| .pytype/ | ||
|
|
||
| # Cython debug symbols | ||
| cython_debug/ | ||
|
|
||
| # PyCharm | ||
| # JetBrains specific template is maintained in a separate JetBrains.gitignore that can | ||
| # be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore | ||
| # and can be added to the global gitignore or merged into this file. For a more nuclear | ||
| # option (not recommended) you can uncomment the following to ignore the entire idea folder. | ||
| #.idea/ |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,28 @@ | ||
| BSD 3-Clause License | ||
|
|
||
| Copyright (c) 2024, Data Science & Global Change Ecology | ||
|
|
||
| Redistribution and use in source and binary forms, with or without | ||
| modification, are permitted provided that the following conditions are met: | ||
|
|
||
| 1. Redistributions of source code must retain the above copyright notice, this | ||
| list of conditions and the following disclaimer. | ||
|
|
||
| 2. Redistributions in binary form must reproduce the above copyright notice, | ||
| this list of conditions and the following disclaimer in the documentation | ||
| and/or other materials provided with the distribution. | ||
|
|
||
| 3. Neither the name of the copyright holder nor the names of its | ||
| contributors may be used to endorse or promote products derived from | ||
| this software without specific prior written permission. | ||
|
|
||
| THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" | ||
| AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE | ||
| IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE | ||
| DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE | ||
| FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL | ||
| DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR | ||
| SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER | ||
| CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, | ||
| OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE | ||
| OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,43 @@ | ||
| # :fish: Fish Module | ||
|
|
||
| <!-- EDIT with your badge link --> | ||
| [](https://github.com/espm-157/climate-python-template/actions/workflows/main.yml) | ||
|
|
||
| ## Team Members | ||
|
|
||
| 🦸 | ||
| 🦹 | ||
|
|
||
| ## 🎓 Learning Objectives | ||
|
|
||
| :octocat: Use of GitHub | ||
| :snake: Use of Jupyter Notebooks | ||
| :abcd: Accessing tabular data | ||
| 📈 Data visualization | ||
| 🌡️Become familiar with data on global climate change | ||
|
|
||
|
|
||
|
|
||
| ## 📖 Content Overview | ||
|
|
||
| [💻 Assignment template](https://github.com/espm-157/climate-python-template/blob/main/notebook.ipynb) | ||
| [💯 Assignment rubric](rubric.md) | ||
|
|
||
| This module will focus on examining a crucial global issue and important scientific debate about the state of global fisheries. In this module we will seek to reproduce some of the most widely cited examples of species collapse ever, and examine the evidence behind an influential and widely cited paper on global fisheries, [Worm et al 2006](http://doi.org/10.1126/science.1132294). However, rather than use the limited data available to Boris Worm and colleagues in 2006, we will be drawing from the best and most recent stock asssement data available today to see how those patterns have faired. | ||
|
|
||
| In this module we will also begin to master one of the most important concepts in data science: manipulation of tabular data using relational database concepts. Instead of working with independent data.frames, we will be working with a large relational database which contains many different tables of different sizes and shapes, but that all all related to each other through a series of different ids. | ||
|
|
||
| ## The Database | ||
|
|
||
| We will use data from the RAM Legacy Stock Assessment Database. In order to better introduce some important emerging technologies, we will be accessing these data directly from a relatively new platform that is now playing a key role in data sharing in machine learning communities, with the memorable name, HuggingFace. We will be streaming data from <https://huggingface.co/datasets/cboettig/ram_fisheries/tree/main/v4.65>. We will have more to say about this approach as we progress. | ||
|
|
||
|
|
||
| ## Science Introduction | ||
|
|
||
| Background abbreviated documentary, features many of the leading authors on both sides https://vimeo.com/44104959 | ||
|
|
||
| ## Links | ||
|
|
||
| [🌐 Course Website](https://espm-157.carlboettiger.info/) | ||
|
|
||
|
|
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,114 @@ | ||
| { | ||
| "cells": [ | ||
| { | ||
| "cell_type": "markdown", | ||
| "id": "95a7abd0", | ||
| "metadata": {}, | ||
| "source": [ | ||
| "# Unit 2: Fisheries Collapse Module Overview\n", | ||
| "\n", | ||
| "This module will focus on examining a crucial global issue and important scientific debate about the state of global fisheries. In this module we will seek to reproduce some of the most widely cited examples of species collapse ever, and examine the evidence behind an influential and widely cited paper on global fisheries, [Worm et al 2006](http://doi.org/10.1126/science.1132294). However, rather than use the limited data available to Boris Worm and colleagues in 2006, we will be drawing from the best and most recent stock asssement data available today to see how those patterns have faired. \n", | ||
| "\n", | ||
| "In this module we will also begin to master one of the most important concepts in data science: manipulation of tabular data using relational database concepts. Instead of working with independent data.frames, we will be working with a large relational database which contains many different tables of different sizes and shapes, but that all all related to each other through a series of different ids. \n", | ||
| "\n", | ||
| "## The Database\n", | ||
| "\n", | ||
| "We will use data from the RAM Legacy Stock Assessment Database. In order to better introduce some important emerging technologies, we will be accessing these data directly from a relatively new platform that is now playing a key role in data sharing in machine learning communities, with the memorable name, HuggingFace. We will be streaming data from <https://huggingface.co/datasets/cboettig/ram_fisheries/tree/main/v4.65>. We will have more to say about this approach as we progress.\n", | ||
| "\n", | ||
| "## Researcher Spotlight: Daniel Pauly\n", | ||
| "\n", | ||
| "Science is done by real people. There are many influential and colorful characters in the global fisheries debate. I want to highlight Professor Pauly not just because he is so famous, but as an early believer in Open Science and Data Science, before we had either of those words. His contributions in making fisheries data more open were ground breaking for their time. I'm also indebted to Professor Pauly whom I had the privilege to meet when I was a junior scientist who had only recently released one of my first software packages, aimed at making data from FishBase more accessible. Academic researchers are typically defined by scientific publications, not software, so I was shocked that Pauly already knew of my software package, and that he encouraged me to continue developing software. Even today that is not common advice, but I believed him, and it's probably a good reason I am where I am today. Scientific textbooks and courses are often critiqued for failing to recognize the contributions of those from minority backgrounds, but as the texts are written on global change ecology, I think none will omit the works for Professor Pauly.\n", | ||
| "\n", | ||
| "\n", | ||
| "## Science Introduction\n", | ||
| "\n", | ||
| "Background abbreviated documentary, features many of the leading authors on both sides https://vimeo.com/44104959" | ||
| ] | ||
| }, | ||
| { | ||
| "cell_type": "code", | ||
| "execution_count": 3, | ||
| "id": "ffb87442", | ||
| "metadata": {}, | ||
| "outputs": [], | ||
| "source": [ | ||
| "import ibis\n", | ||
| "from ibis import _\n", | ||
| "import ibis.selectors as s\n", | ||
| "import seaborn.objects as so" | ||
| ] | ||
| }, | ||
| { | ||
| "cell_type": "markdown", | ||
| "id": "cdbc9dc1-89db-4190-9683-9b7833e64207", | ||
| "metadata": {}, | ||
| "source": [ | ||
| "\n", | ||
| "# Exercise 1: Investigating the North-Atlantic Cod\n", | ||
| "\n", | ||
| "Now we are ready to dive into our data. First, We seek to replicate the following figure from the Millennium Ecosystem Assessment Project using the RAM data.\n", | ||
| "\n", | ||
| "\n" | ||
| ] | ||
| }, | ||
| { | ||
| "cell_type": "code", | ||
| "execution_count": null, | ||
| "id": "c8a426ad-d13a-4011-83f0-108e09036853", | ||
| "metadata": {}, | ||
| "outputs": [], | ||
| "source": [] | ||
| }, | ||
| { | ||
| "cell_type": "markdown", | ||
| "id": "88f42ebf-a09d-4cf9-8748-12bf7889c2db", | ||
| "metadata": {}, | ||
| "source": [ | ||
| "# Excersise 2: Global Fisheries \n", | ||
| "\n", | ||
| "## Stock Collapses\n", | ||
| "\n", | ||
| "We seek to replicate the temporal trend in stock declines shown in [Worm et al 2006](http://doi.org/10.1126/science.1132294):\n", | ||
| "\n", | ||
| "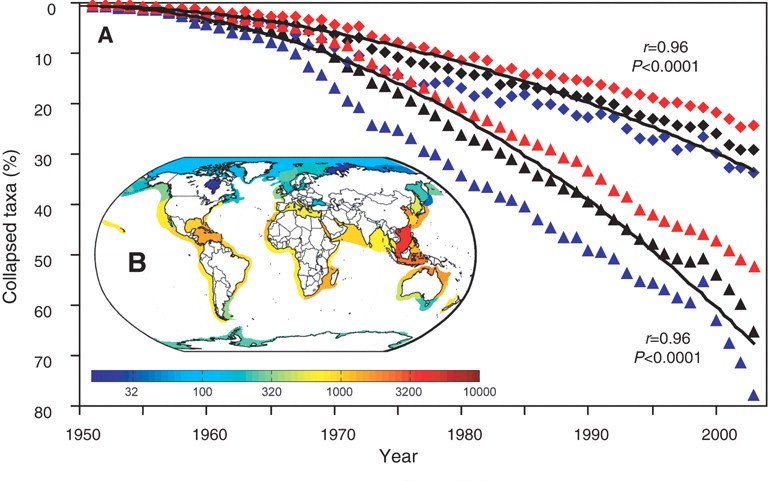" | ||
| ] | ||
| }, | ||
| { | ||
| "cell_type": "code", | ||
| "execution_count": null, | ||
| "id": "0cf887f4-6d94-45f4-8667-512b302d65c6", | ||
| "metadata": {}, | ||
| "outputs": [], | ||
| "source": [] | ||
| } | ||
| ], | ||
| "metadata": { | ||
| "jupytext": { | ||
| "formats": "md:myst,ipynb" | ||
| }, | ||
| "kernelspec": { | ||
| "display_name": "Python 3 (ipykernel)", | ||
| "language": "python", | ||
| "name": "python3" | ||
| }, | ||
| "language_info": { | ||
| "codemirror_mode": { | ||
| "name": "ipython", | ||
| "version": 3 | ||
| }, | ||
| "file_extension": ".py", | ||
| "mimetype": "text/x-python", | ||
| "name": "python", | ||
| "nbconvert_exporter": "python", | ||
| "pygments_lexer": "ipython3", | ||
| "version": "3.10.12" | ||
| }, | ||
| "vscode": { | ||
| "interpreter": { | ||
| "hash": "31f2aee4e71d21fbe5cf8b01ff0e069b9275f58929596ceb00d14d90e3e16cd6" | ||
| } | ||
| } | ||
| }, | ||
| "nbformat": 4, | ||
| "nbformat_minor": 5 | ||
| } |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,4 @@ | ||
| pandas | ||
| seaborn | ||
| ibis-framework[duckdb] | ||
| nbval |
Oops, something went wrong.