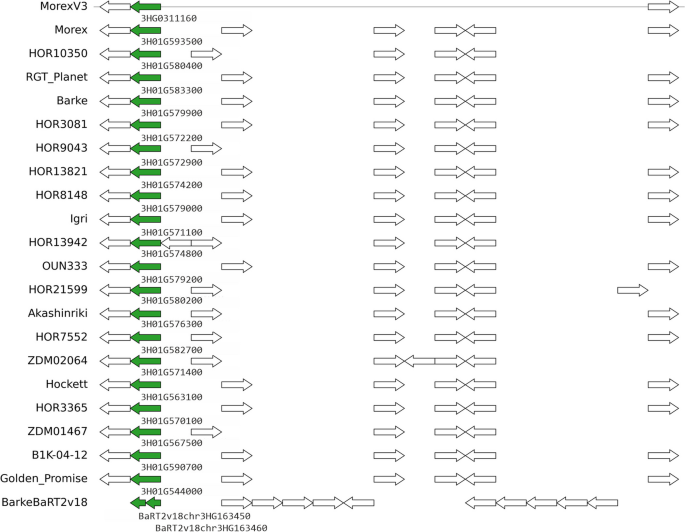
Pangenes are clusters of gene models/alleles/haplotypes found in barley assemblies in a similar genomic location.
You can browse pangenes and display multiple sequence alignments (MSA) of encoded proteins at https://eead-csic-compbio.github.io/barley_pangenes.
Also, MorexV3 HC genes in maps produced by BARLEYMAP link out to these MSAs.
The pangene matrices pangene_matrix.tr.bed and pangene_matrix_genes.tr.tab in BED and TAB format describe clusters of gene models found to be collinear across barley assemblies and annotations in the order:
MorexV3 Morex Barke HOR10350 HOR3081 HOR9043 OUN333 HOR7552 Igri HOR21599 HOR13942 Akashinriki HOR8148 RGT_Planet HOR13821 B1K-04-12 HOR3365 Hockett ZDM02064 ZDM01467 Golden_Promise BarkeBaRT2v18
The TAB file contains one cluster per row, the 1st column being the cluster name.
The BED file describes the same clusters with the first four columns indicating the position of the relevant gene model in the MorexV3 reference and the cluster name. Clusters that do not include MorexV3 genes do not have an exact position, they are placed within an interval defined by two MorexV3 genes. Those rows start with '#'.
Input genome assemblies (FASTA) and gene annotations (GFF) were obtained from IPK, Ensembl Plants and JHI as explained in https://doi.org/10.1186/s13059-023-03071-z. These files are available at https://zenodo.org/records/7961646.
Barley pangenes were produced by computing Whole Genome Alignments followed by gene model overlap and clustering with get_pangenes.pl version 04102024 with the following arguments:
plant-scripts/pangenes/get_pangenes.pl -d barley -s '^chr\d+H' -m cluster -r MorexV3 -H -t 0 > log.barley.H.t0.MorexV3.txt
Note this approach uses nucleotide sequences, not protein sequences, being an alaternative to protein-based orthogroups in https://panbarlex.ipk-gatersleben.de.
Multiple alignments of resulting .cds.faa clusters were computed with clustal-omega v1.2.4.
To reduce avoid annotation bias and redundancy, BaRTv2 and MorexV3 gene models were removed to carry out haplotype analysis of trimmed protein sequences, which was performed as explained in https://github.com/Ensembl/plant-scripts/tree/master/pangenes#example-6-estimation-of-haplotype-diversity and summarized on poster http://hdl.handle.net/10261/368616. This was done with v10012024 data.
Protocol first published at:
Contreras-Moreira B, Saraf S, Naamati G et al (2023) GET_PANGENES: calling pangenes from plant genome alignments confirms presence-absence variation. Genome Biol 24, 223. https://doi.org/10.1186/s13059-023-03071-z
Haplotype diversity poster:
Contreras-Moreira B, Montardit F, Sàrria J, Igartua E, Casas AM (2024) Analysis of collinear genes across the barley pangenome reveals that 85% of protein coding genes have two or more alleles. 1er Congreso de la Sociedad Española de Bioinformática y Biología Computacional (SEBiBC), C-0314. http://hdl.handle.net/10261/368616
Multiple sequence alignments produced with clustal-omega v1.2.4:
Sievers F, Wilm A, Dineen D, et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038%2Fmsb.2011.75
Annotated genomes from:
Jayakodi, M., Padmarasu, S., Haberer, G. et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588, 284–289 (2020). https://doi.org/10.1038/s41586-020-2947-8
Mascher M, Wicker T, Jenkins J, et al. Long-read sequence assembly: a technical evaluation in barley, The Plant Cell, Volume 33, Issue 6, June 2021, Pages 1888–1906, https://doi.org/10.1093/plcell/koab077
Coulter M, Entizne JC, Guo W, et al. BaRTv2: a highly resolved barley reference transcriptome for accurate transcript-specific RNA-seq quantification. Plant J. 2022 111(4):1183-1202. https://doi.org/10.1111%2Ftpj.15871
[PRIMA GENDIBAR, PCI2019-103526] supported by Horizon 2020
[A08_23R] funded by Aragón goverment
[FAS2022_052, INFRA24018, conexión BCB] funded by CSIC
[PID2022-142116OB-I00] by MICIU/AEI/10.13039/501100011033