Lydia Carroll, Benjamin Hoertnagl-Pereira, Ryan Walter
JHED: lcarro12, bhoertn1, rwalte25
lcarro12 @jhu.edu, [email protected], [email protected]
600.226.01 | CS226 Data Structures
Project 4 - Image Segmentation
Files:
Distance.java
GVertex.java
P4C.java
Partition.java
Pixel.java
PixelDistance.java
PQHeap.java
QueueEmptyException.java
StoreInfo.java
WEdge.java
WGraphP4.java
WGraphP4Test.java
*--------------------------------------------------------------------------------*
Command line needed to run program:
java P4C [pic_file_name].png [k_value]
Picture files passed to program should be of type png. Kvalue is a tuning variable and is
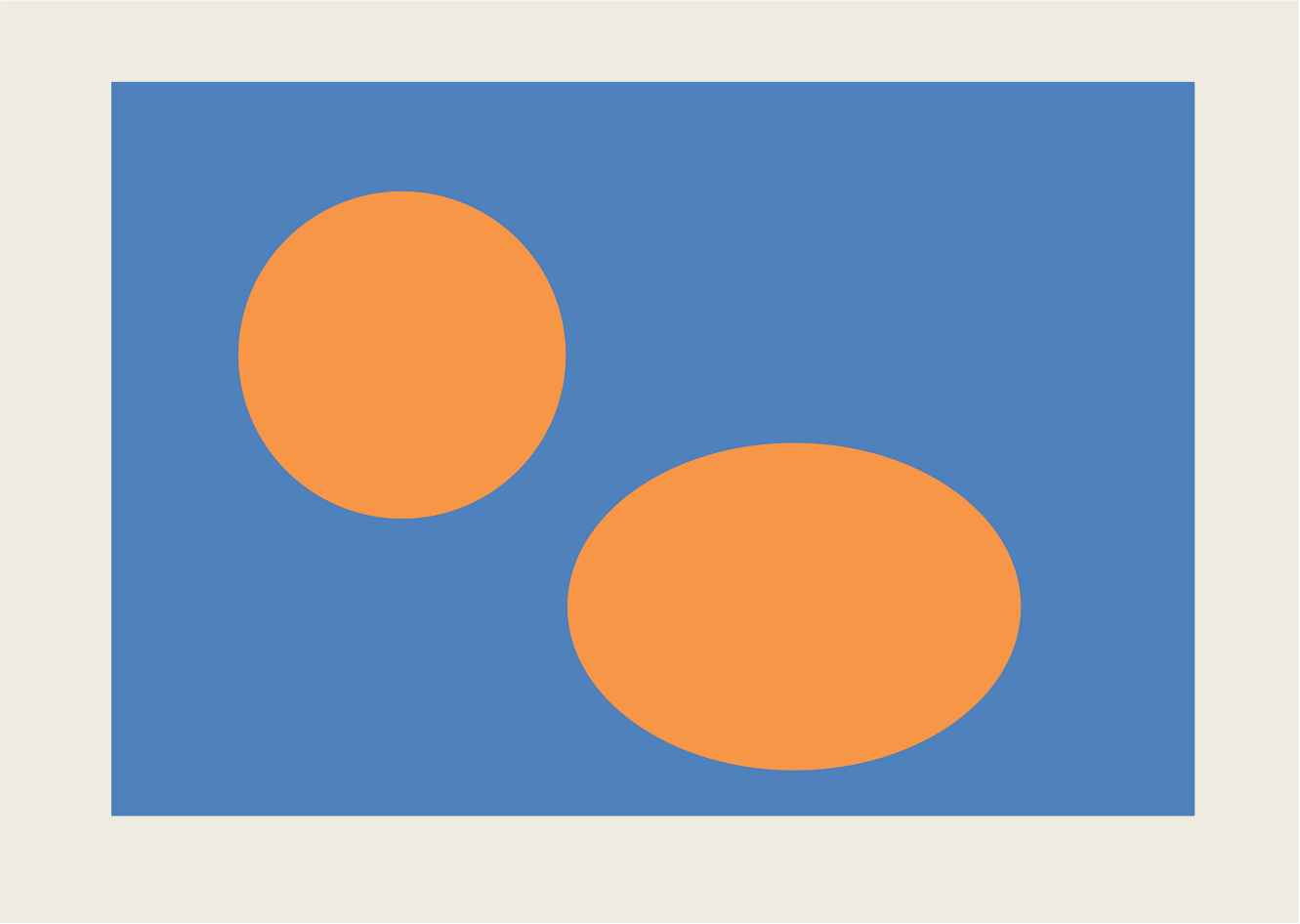
chosen by user—-results will vary depending on K. Usually K ~100 works. Test4 picture as
input (blue background, orange circles/ ovals) works beautifully at K=100.
*--------------------------------------------------------------------------------*
Discussion
Our original graph implementation was fairly easy to integrate into Part C. Because we
stored a list of neighboring vertices and incident edges in each vertex, it was easy to
go from the list of edges returned by segementer() to a graph, then from a graph to a
depth-first search. Our greatest challenge for this part was writing efficiently to reduce
run time. We ultimately were able to progress from quadratic time to near-constant time by
changing from storing lists of vertices in segmenter() to only storing the minimum, maximum
and size of each partition. After this change our solution was slowest at the depth-first
search, which we were not sure how to make faster. We tested k-values from 20 to 1000 and
were happiest with a k-value of 100. It worked particularly well for the given input file
test4.png and produced nicely rounded edges. Our function runs in under 7 seconds for the
largest test file that we were given so we are happy with our time complexity.
In this project, we have seen how to transform a problem into a graph representation, solve it as a graph, and then transform it back again. The starting point for this project is work by Pedro Felzenszwalb and Dan Huttenlocher on provable algorithms for image segmentation: http://cs.brown.edu/~pff/papers/seg-ijcv.pdf The idea in this paper is to reduce image segmentation to the problem of producing a minimal spanning forest.
This paper has been used in many forms, including, for example, AR Tag recognition: https://april.eecs.umich.edu/pdfs/olson2011tags.pdf
This project solves 3 problems:
- Flesh out a graph implementation starting with an existing solution we will provide.
- Implement a minimally weighted spanning tree algorithm using Kruskal's Algorithm.
- Adapt this algorithm to image segmentation as we describe below and apply the segmentation algorithm to real images.
For this project, we chose to implement the graph abstraction of images via an adjacency list due to its superior performance in comparison to an edge list or adjacency matrix. We began by writing a generic vertex class, GVertex.java, to represent nodes of each graph. Given that for our specific application, the graph representation of an image would be made once and would not need to support removal and addition functionality, we made the design choice to optimize find and adjacency functions by storing the list of adjecent vertices in each vertex, along with the list of edges as expected.Next, we created an weighted edge class, WEdge.java, to represent links between nodes in the graph. This edge class implements the Comparable interface, comparing edges by their weights. The graph implementation itself, WGraphP4.java, implements the Weighted Graph ADT interface in the form of WGraph.java.
We decided to use the JUnit framework to create a testing suite for our graph implementation, and the vertex and edge clases that it ecnompasses. This can be found in the WGraphP4Test.java file.
Using our graph representation, we implemented Kruskal's algorithm to find the minimum spanning tree or forest of a graph, depending on if the graph was connected or not. This algorithm makes use of a priority queue of edges based on their weights as keys, and fast disjoint set union.
To implement a Priority Queue ADT, we chose to use an array representation of a minimum binary heap, PQHeap.java. The heap supports a linear time bottom up build to ensure peak efficiency. From here, we use the priority queue to sort the edges of a given graph in increasing order, and build the graph up by adding each edge one at a time and checking for cycles, until all vertices were included - thus, a spanning tree/forest.
Again, for testing purposes, we created a JUnit testing suit for the prioirty queue, PQHeapTest.java.
Now, we were able to create a graph representation of an image.
First, we created graph node representation for each pixel. We read in an image using the BufferedImage Java API to represent the given image as a two dimensional grid of color values. For the purposes of this project, we took the neighborhood of a pixel to be its right, left, up and down neighbors. We created the respectve edges between nodes, with the weight being the square root of the sume of each difference in color value (RGB).
From here, we created a variation on Kruskal's algorithm that includes an additional condition on merges (partition unions) -- namely it checks the condition:
Diff(A u B) <= Min(Diff(A), Diff(B)) + K/(|A| + |B|)
and only allows the merge if this condition is true. Diff(A) is a tuple which represents the difference between the minimum and maximum of each component (byte) of all the pixel values in the graph component A, |A| means the size (number of vertices) in A, and K is a constant explained below.
Note that K is a "tuning parameter." You should now write a new version of Kruskal called Segmenter. Segmenter is identical to Kruskal except that when you get to the point of merging two spanning components, if the condition is not satisfied, you do not do the merge. The result of this new algorithm will be a spanning forest represented as a list of edges. Each connected component of the forest will be a portion of the original image that has a consistent appearance.
We now found the connected components of the spanning forest by running a modified depth first search on all the vertices in the edge list returned by our segmenter function. For each set of pixels in the given forest, we created a new image representaion and wrote it out as a new file. We later added a constaint that required a certain number of pixes to be present in a forrest (in relation to the total number of pixels of an image), so that anomalies were not considered and only the cleanest of segmented images were kept.
*--------------------------------------------------------------------------------*
Part A
Summary : Our greatest challenge for Part A was understanding the Java syntax to implement
generics. Once this was understood we proceeded rapidly.
GVertex.java - Lydia
There were early problems with generics and implementing comparable which were quickly
resolved. Each instance of GVertex holds a holds a list of neighboring vertices as well
as a list of incident edges.
GVertexTest.java - Ben and Lydia
It tests GVertex by comparing the results of its methods to the results of the Vertex methods.
WEdge.java - Ryan and Ben
Early problems with generics were resolved as Part B was written.
WGraphP4.java - Ben, everything but Kruskal’s
The given graph implementation using an adjacency matrix was modified to work with the
adjacency lists present in GVertex. Adjacency lists were chosen because we thought they
would be easiest to implement down the line despite the additional upfront bookkeeping. The
graph was implemented to best support insertion operations of vertices and edges at the expense
of making removal operations on these objects more expenseive. This design choice was made
because our appication did not utilize removal operations, so we wanted to make sure the core
operations used were as fast as possible.
*--------------------------------------------------------------------------------*
Part B
PQHeap.java - Ryan
This priority queue implementation uses the Priority Queue interface given in the assignment.
Creating the Comparator constructor and default constructor was initially difficult and somewhat
unclear. The PQHeap is represented via an ArrayList for a bottom-up construction.
PQHeapTest.java - Ryan
Extensively tests the written PQHeap implementation.
WGraphP4.java - Lydia, Kruskal’s
This algorithm returns a spanning forest of edges for an input graph. It was not difficult to
implement thanks to the excellent slides given in class.
WGraphP4Test.java - Ben and Lydia
We extensively tested our graph implementation before moving on to Part C. We first wrote our
own tests then looked at the tests given by Prof. Selinski and added them.
*--------------------------------------------------------------------------------*
Part C
Distance.java - N/A
This interface was given for the color difference between pixels in the starter code.
P4C.java
- Lydia, segmenter, diff
- Ben, readImage, writeImage, and depth-first search
- Ryan, readImage, writeImage, and depth-first search
Initially we were confused because the depth-first search of the segmented output was only
showing 7 edges, but then we realized we were accidentally resetting all edges every call in
the graph construction loop. Once we fixed this the main function ran and worked but was
exceptionally slow. To speed it up, we switched from storing lists of vertices in segmenter
to only storing the minimum, maximum and size of each partition. We then improved our output
images by excluding any subtrees of less than 50 vertices. This eliminated the results of
over-segmentation.
Partition.java - N/A
This class tracks which vertices are in the same cloud. It was given in the starter code.
Pixel.java - Ben
This class stores the color, row and column values of a pixel in an image.
PixelDistance.java - Ben
This implements the given Distance interface to find the color difference between pixels.
QueueEmptyException.java - Ryan
This exception is thrown by PQHeap if remove() is attempted when it is empty.
StoreInfo.java - Ryan and Lydia
This class holds the minimum, maximum, and number of vertices in a given partition. It is
used to calculate the difference between partitions used in segmenter.