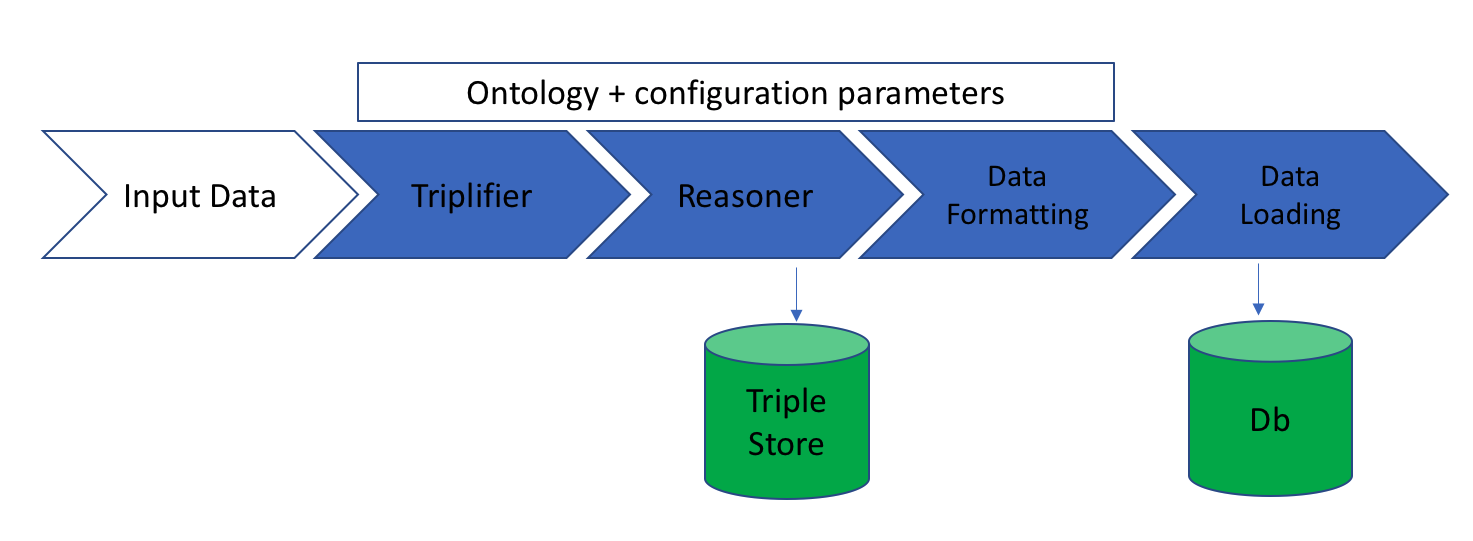
A flexible, scalable pipeline for integration and alignment of multiple data sources. The code is written to be adaptable to all kinds of data, ontologies (OWL), or reasoning profiles, and output is compatible with any type of storage technology.
A good way to start with the ontology-data-pipeline is to fork or clone one of the applications which use this code. This includes:
- fovt-data-pipeline a data pipeline for processing vertebrate trait measurements
- ppo-data-pipeline a data pipeline for processing plant phenology observations
Step 1: Install docker
Step 2: Run the application. On the commandline, you can execute the script like so:
# make sure you have the latest docker container
docker pull jdeck88/ontology-data-pipeline
# run the pipeline help in the docker container
docker run -v "$(pwd)":/process -w=/app -ti jdeck88/ontology-data-pipeline python pipeline.py -h
usage: pipeline.py [-h] [--drop_invalid] [--log_file]
[--reasoner_config REASONER_CONFIG] [-v] [-c CHUNK_SIZE]
[--num_processes NUM_PROCESSES]
data_file output_dir ontology config_dir
ontology data pipeline command line application.
positional arguments:
data_file Specify the data file to load.
output_dir path of the directory to place the processed data
ontology specify a filepath/url of the ontology to use for
reasoning/triplifying
config_dir Specify the path of the directory containing the
configuration files.
optional arguments:
-h, --help show this help message and exit
--drop_invalid Drop any data that does not pass validation, log the
results, and continue the process
--log_file log all output to a log.txt file in the output_dir.
default is to log output to the console
--reasoner_config REASONER_CONFIG
optionally specify the reasoner configuration file.
Default is to look for reasoner.config in the
configuration directory
-v, --verbose verbose logging output
-c CHUNK_SIZE, --chunk_size CHUNK_SIZE
chunk size to use when processing data. optimal
chunk_size for datasets with less then 200000
recordscan be determined with: num_records / num_cpus
--num_processes NUM_PROCESSES
number of process to use for parallel processing of
data. Defaults to cpu_count of the machine
As an alternative to the commandline, params can be placed in a file, one per
line, and specified on the commandline like 'pipeline.py @params.conf'.
The ontology-data-pipeline operates on a set of configuration files, which you can specify in the configuration directory.
The following text describes the operation of the pipeline and the steps involved.
-
Triplifier
This step provides provides basic data validation and generates the RDF triples, assuming validation passes, needed for the reasoning phase. Each project will need to contain a
configdirectory with the following files that will be used to triplify the preprocessed data:NOTE: Wherever there is a uri expressed in any of the following files, you have the option of using ontology label substitution. If the uri is of the format
{label name here}, the appropriate uri will be substituted from the provided ontology. See the ROBOT for details term identifier abbreviations. -
Reasoning
This step uses the ROBOT project to perform reasoning on the triplified data in the triplifier step, in conjunction with logic contained in the provided ontology. An example of the use of the ROBOT file is given below, calling the robot.jar which is in the
/libdirectory. Please note that the pipeline code calls this command for you and normally you do not need to call this command directly. Here we are illustrating a direct-use of the robot command if you want to explore how this works within the ontology-data-pipeline environment.
java -jar lib/robot.jar reason -r elk \
--axiom-generators "InverseObjectProperties ClassAssertion" \
-i sample_data/unreasoned_data.ttl \
--include-indirect true \
--exclude-tautologies structural \
reduce \
-o sample_data/reasoned_data.ttl
In the above example, the reasoner use is ELK, with InverseObjectProperties and ClassAssertion axioms specified. The input file is sample_data/unreasoned_data.ttl. We tell the reasoning engine to include indirect inferences, which lets us assert recursive SubClass relationships. Exclude tautoligies tells the reasoner to not include assertions which will always be true. The reduce command eliminates redundant assertions. Finally, the output file is sample_data/reasoned_data.ttl. The most critical step of the reasoning process is including the indirect inferences: this is the step that lets us export our end-results to a simple Document Store and not rely on "smart" applications which are able to iterate recursive relationships. Examples included in the test directory.
-
Data Formatting
This step takes a custom SPARQL query and generates csv files for each file outputted in the Reasoning step using ROBOT. You must write a SPARQL query yourself to format data as you wish it to appear. You can refer to the example below for a query that turns the sample input (reasoned) file into CSV. If no sparql query is found, then this step is skipped. An example of the data Formatting step is given below calling the robot.jar which is in the
/libdirectory. Please note that the pipeline code calls this command for you and normally you do not need to call this command directly. Here we are illustrating a direct-use of the robot command if you want to explore how this works within the ontology-data-pipeline environment.
java -jar lib/robot.jar query --input sample_data/reasoned_data.ttl \
--query sample_data/fetch_reasoned.sparql \
sample_data/reasoned_data.ttl.csv
In the above example, we call the ROBOT query sub-command and give an input file of sample_data/reasoned_data.ttl (this is the output of the above command), and tell it to use the sample_data/fetch_reasoned.sparql sparql command as a guide to produce the output file sample_data/reasoned_data.ttl.csv.
Project configuration files include entity.csv, mapping.csv, relations.csv, and any files defining controlled vocabularies that we want to map rdf:types to. The remaining configuration files below are found in the config directory. Together, these are the required configuration files we use for reasoning against the application ontology (e.g. Plant Phenology Ontology). These files configure the data validation, triplifying, reasoning, and rdf2csv converting.
The following files are required:
-
entity.csv(found in each project directory) - This file specifies the entities (instances of classes) to create when triplifying. The file expects the following columns:-
aliasThe name used to refer to the entity. This is usually a shortened version of the class label.
-
concept_uriThe uri which defines this entity (class).
-
unique_keyThe column name that is used to uniquely identify the entity. Whenever there is a unique value for the property specified by "unique key", a new instance will be created. e.g. "record_id"
-
identifier_rootThe identifier root for each unique entity (instance created). E.g. urn:observingprocess/ would be the root of urn:observingprocess/record1
-
-
mapping.csv(found in each project directory)-
columnThe name of the column in the csv file to be used for triplifying
-
uriThe uri which defines this column. These generally are data properties.
-
entity_aliasThe alias of the entity (from entity.csv) this column is a property of
-
-
relations.csv(found in each project directory)-
subject_entity_aliasThe alias of the entity which is the subject of this relationship
-
predicateThe uri which defines the relationship
-
object_entity_aliasThe alias of the entity which is the object of this relationship
-
The following files are optional:
-
rules.csv- This file is used to setup basic validation rules for the data. The file expects the following columns:-
ruleThe name of the validation rule to apply. See rule types below. Note: a default
ControlledVocabularyrule will be applied to thephenophase_namecolumn for the names found in the phenophase_descriptions.csv file -
columnsPipe
|delimited list of columns to apply the rule to -
levelEither
WARNINGorERROR.ERRORwill terminate the program after validation.WARNINGSwill be logged. Case-Insensitive. Defaults toWARNING -
listOnly applicable for
ControlledVocabularyrules. This refers to the name of the file that contains the list of the controlled vocab
RequiredValue- Specifies columns which can not be emptyUniqueValue- Checks that the values in a column are uniqueControlledVocabulary- Checks columns against a list of controlled vocabulary. The name of the list is specified in thelistcolumn inrules.csvInteger- Checks that all values are integers. Will coerce values to integers if possibleFloat- Checks that all values are floating point numbers (ex. 1.00). Will coerce values to floats if possible
-
-
Any file specified in
rules.csvlistcolumn is required. The file expects the following columns:field- Specifies a valid value. This is the values expected in the input data filedefined_by- Optional value which will replace the field when writing triples
-
fetch_reasoned.sparql- Sparql query used to convert reasoned data to csv
The ontology-data-pipeline is designed to be run as a Docker container. However, you can also run the codebase from sources by checking out this repository and following the instructions at python instructions. Information on building the docker container is contained at docker instructions.