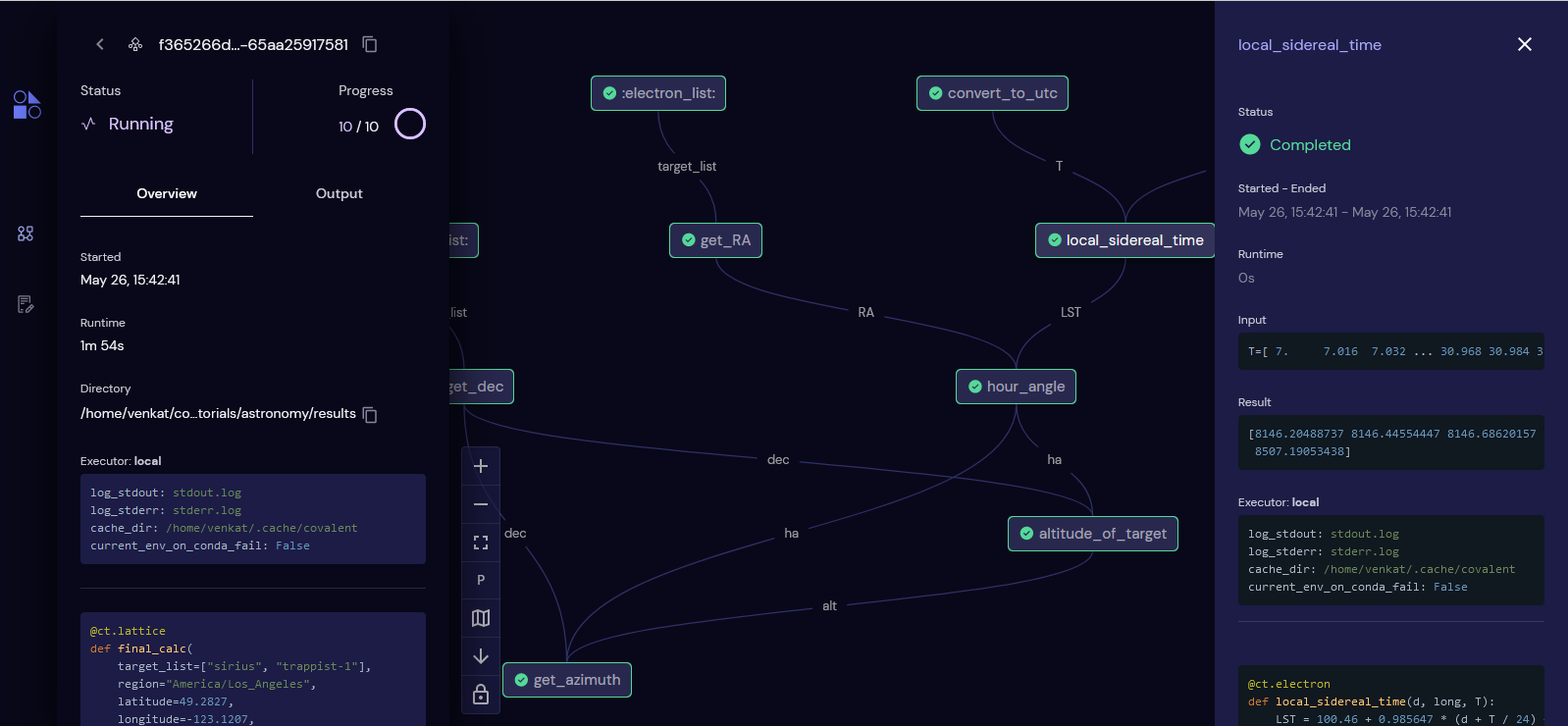
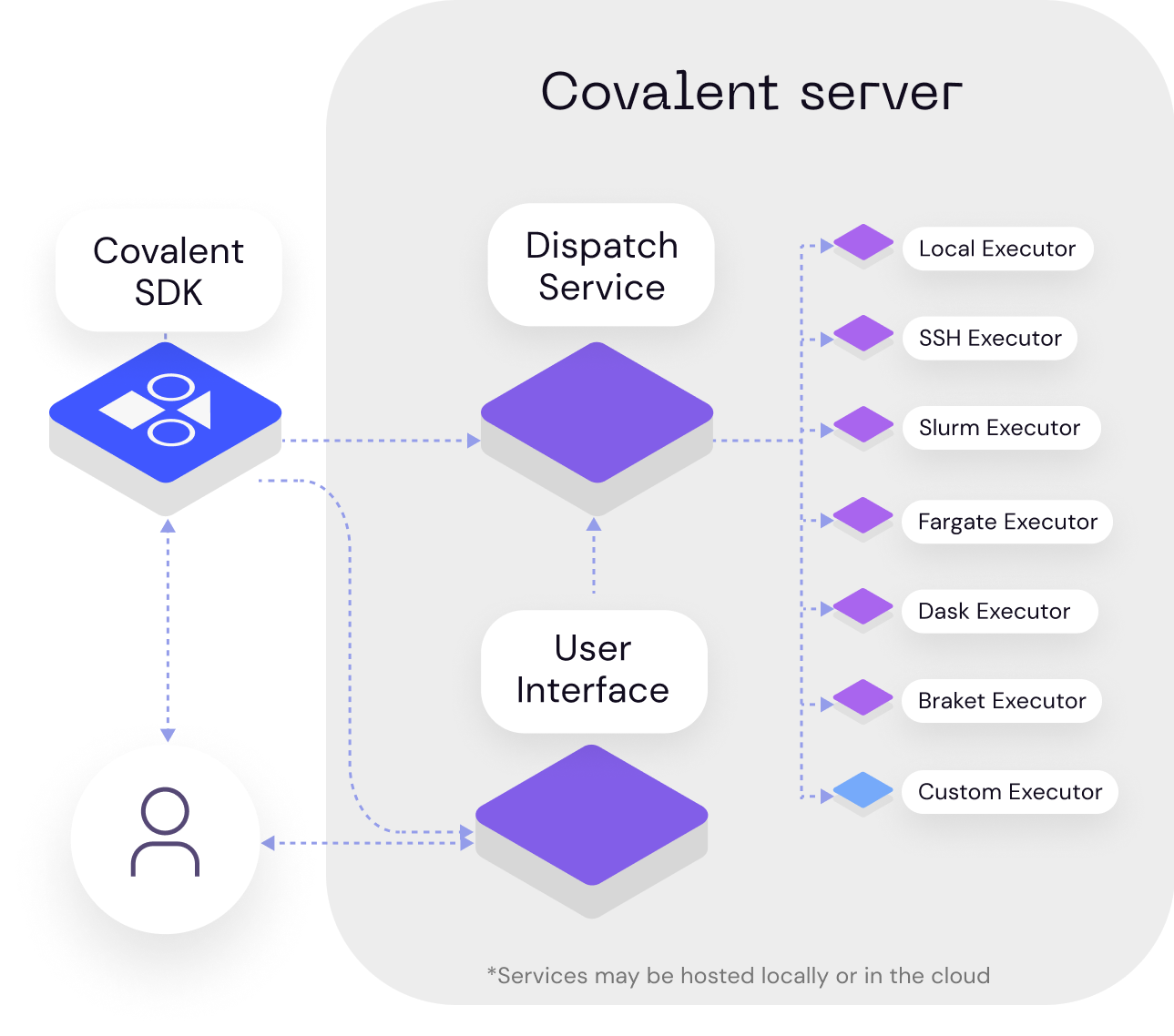
Covalent is a Pythonic workflow tool used to execute HPC and quantum tasks in heterogenous environments. Computational scientists and engineers use Covalent to...
- rapidly iterate prototypes and exploratory research models
- automate, manage, and share reproducible experiments
- visualize data and task dependencies in an interactive user interface
- run code in heterogenous compute environments, including in hybrid-cloud and hybrid-quantum configurations
- understand where time and money is spent across a project
Covalent may be deployed locally or on a remote server. Covalent is rapidly expanding to include support for a variety of cloud interfaces, including HPC infrastructure tools developed by major cloud providers and emerging quantum APIs. It has never been easier to deploy your code on the world's most advanced computing hardware with Covalent.
Read more in the official documentation.
- Purely Pythonic: No need to learn any new syntax or mess around with YAML. Construct your complex workflow programmatically with native Python functions. By just adding one-line decorators to your functions, you can supercharge your experiments.
- Native parallelization: Covalent natively parallelizes mutually independent parts of your workflow.
- Monitor with UI: Covalent provides an intuitive and aesthetically beautiful browser-based user interface to monitor and manage your workflows.
- Abstracted dataflow: No need to worry about the details of the underlying data structures. Covalent takes care of data dependencies in the background while you concentrate on understanding the big picture.
- Result management: Covalent manages the results of your workflows. Whenever you need to modify parts of your workflow, from inputs to components, Covalent stores and saves the run of every experiment in a reproducible format.
- Little-to-no overhead: Covalent is designed to be as lightweight as possible and is optimized for the most common use cases. Covalent's overhead is less than 0.1% of the total runtime for typical high compute applications and often has a constant overhead of ~ 10-100μs -- and this is constantly being optimized.
- Interactive: Unlike other workflow tools, Covalent is interactive. You can view, modify, and re-submit workflows directly within a Jupyter notebook.
For a more in-depth description of Covalent's features and how they work, refer to the Concepts page in the documentation.
Begin by starting the Covalent servers:
covalent startNavigate to the user interface at http://localhost:48008 to monitor workflow execution progress.
In your Python code, it's as simple as adding a few decorators! Consider the following example which uses a support vector machine (SVM) to classify types of iris flowers.
| Without Covalent | With Covalent |
|---|---|
from numpy.random import permutation
from sklearn import svm, datasets
def load_data():
iris = datasets.load_iris()
perm = permutation(iris.target.size)
iris.data = iris.data[perm]
iris.target = iris.target[perm]
return iris.data, iris.target
def train_svm(data, C, gamma):
X, y = data
clf = svm.SVC(C=C, gamma=gamma)
clf.fit(X[90:], y[90:])
return clf
def score_svm(data, clf):
X_test, y_test = data
return clf.score(
X_test[:90],
y_test[:90]
)
def run_experiment(C=1.0, gamma=0.7):
data = load_data()
clf = train_svm(
data=data,
C=C,
gamma=gamma
)
score = score_svm(data=data, clf=clf)
return score
result=run_experiment(C=1.0, gamma=0.7) |
from numpy.random import permutation
from sklearn import svm, datasets
import covalent as ct
@ct.electron
def load_data():
iris = datasets.load_iris()
perm = permutation(iris.target.size)
iris.data = iris.data[perm]
iris.target = iris.target[perm]
return iris.data, iris.target
@ct.electron
def train_svm(data, C, gamma):
X, y = data
clf = svm.SVC(C=C, gamma=gamma)
clf.fit(X[90:], y[90:])
return clf
@ct.electron
def score_svm(data, clf):
X_test, y_test = data
return clf.score(
X_test[:90],
y_test[:90]
)
@ct.lattice
def run_experiment(C=1.0, gamma=0.7):
data = load_data()
clf = train_svm(
data=data,
C=C,
gamma=gamma
)
score = score_svm(
data=data,
clf=clf
)
return score
dispatchable_func = ct.dispatch(run_experiment)
dispatch_id = dispatchable_func(
C=1.0,
gamma=0.7
)
result = ct.get_result(dispatch_id) |
>>> print(result)
0.988888888 |
>>> print(f"""
... status = {result.status}
... input = {result.inputs}
... result = {result.result}
... """)
status = Status(STATUS='COMPLETED')
input = {'C': 1.0, 'gamma': 0.7}
result = 0.988888888 |
For more examples, please refer to the Covalent tutorials.
Covalent is developed using Python version 3.8 on Linux and macOS. The easiest way to install Covalent is using the PyPI package manager:
pip install covaRefer to the Getting Started guide for more details on setting up. For a full list of supported platforms, consult the Covalent compatibility matrix. Read this guide if you are migrating from cova version 0.3x.
Users compose workflows using the Covalent SDK and submit them to the Covalent server. Upon receiving a workflow, the server analyzes the dependencies between tasks and dispatches each task to its specified execution backend. Independent tasks may be executed concurrently. The Covalent UI displays the execution progress of each workflow at the level of individual tasks.
The official documentation includes tips on getting started, some high level concepts, a handful of tutorials, and the API documentation. To learn more, please refer to the Covalent documentation.
To contribute to Covalent, refer to the Contribution Guidelines. We use GitHub's issue tracking to manage known issues, bugs, and pull requests. Get started by forking the develop branch and submitting a pull request with your contributions. Improvements to the documentation, including tutorials and how-to guides, are also welcome from the community. Participation in the Covalent community is governed by the Code of Conduct.
The latest release of Covalent is now out and available for community use. It comes with new features including the integration of Dask into Covalent as the default executor/compute backend for workflows. As part of this release, we have also updated the CLI to allow users the flexibility to choose the default compute backend between Dask and Local. The default executor for workflow electrons are then set accordingly at runtime. We have also made it simpler for users to parse the logs generated by Covalent by providing a convenient CLI. A summary of the feature releases is provided below
- Dask local cluster is now the default compute backend for executing workflows
- Covalent now supports the ability to switch between
DaskandLocalexecutors covalent logshas now be added to facilitate parsing logs generated by Covalent
History of changes can be viewed in the Changelog.
Please use the following citation in any publications:
W. J. Cunningham, S. K. Radha, F. Hasan, J. Kanem, S. W. Neagle, and S. Sanand. Covalent. Zenodo, 2022. https://doi.org/10.5281/zenodo.5903364
Covalent is licensed under the GNU Affero GPL 3.0 License. Covalent may be distributed under other licenses upon request. See the LICENSE file or contact the support team for more details.