-
Notifications
You must be signed in to change notification settings - Fork 9
Crossspecies comparisons
One of the great advantages of NHP imaging is that we now have the same (or very similar) data for the NHP and the human, allowing direct like-for-like comparisons. However, this type of comparison is challenging as the brains differ substantially in size, organization, and morphology. This means that a simple registration into a template space, analogous to warping different individual human brains to MNI152, is not possible.
Common feature space approach
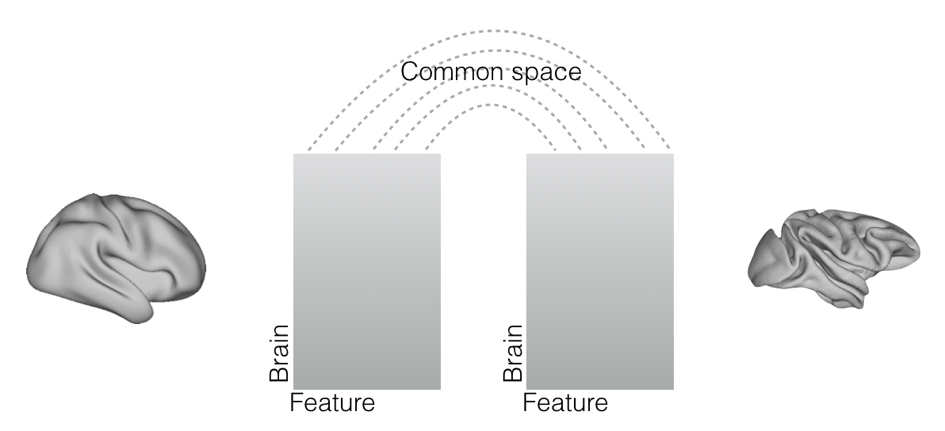
One recent approach is to describe the brains of the different species in an abstract feature space that is common to the species at hand (Mars et al., 2018, eLife; Mars et al., 2018, Curr Opin Behav Sci; Xu et al., 2019, bioRxiv). The idea is that brains can differ in multiple respects, but some of these are often irrelevant to the question at hand. For instance, when studying connections, brain size and sulcal morphology might be unimportant. By describing brain regions then in terms of their profiles of connectivity with homologous areas that are present in both brains or in terms of connectivity with homologous white matter tracts, it is possible to directly compare each brain region in one brain with each brain region in the other (Mars et al., 2016, Neurosci Biobehav Rev; Mars et al., 2018, eLife), without being bothered by the irrelevant features of size and sulcal morphology. In effect, one is now using a common grammar to describe the two brains. One can even take this further and use a data reduction algorithm, for instance, Laplacian eigenmapping, to create a lower-level ‘embedding space’ in which to describe both brains (cf. Xu et al, 2019, bioRxiv).
|
|---|
| Figure 9. Common space approach as set out in Mars et al. (2018, eLife). |
This approach has only been around for a short time, but it has its historical roots in earlier approaches. For instance, Van Essen and Dieker (2007, Neuron) used surface-based registration to align homologous landmarks across species to identify areas of cortical expansion in the human compared to the macaque monkey. A similar approach was taken by Chaplin et al. (2013, J Neurosci).
Another approach is based on brain dynamics. By having humans and NHPs process the same stimulus sequence, it is possible to compare profiles of brain activation over time between the two brains (e.g., Mantini et al., 2012, Nat Methods). In effect, the common space here is the time course of activation.
Tools for implementing such approaches are available. The original Van Essen surface-based registration was implemented in Caret. The common space approach using areal connectivity or white matter tracts, as well as recent extensions (e.g., Eichert et al., 2020, eLife), are implemented in Mr Cat. There are a number of toolboxes available for gradient processing (Haak et al., 2018, NeuroImage; Vos de Waal et al., 2020, Comm Biol) and its use in between-species embedding space is available here.
Brain matching via homologous sulci
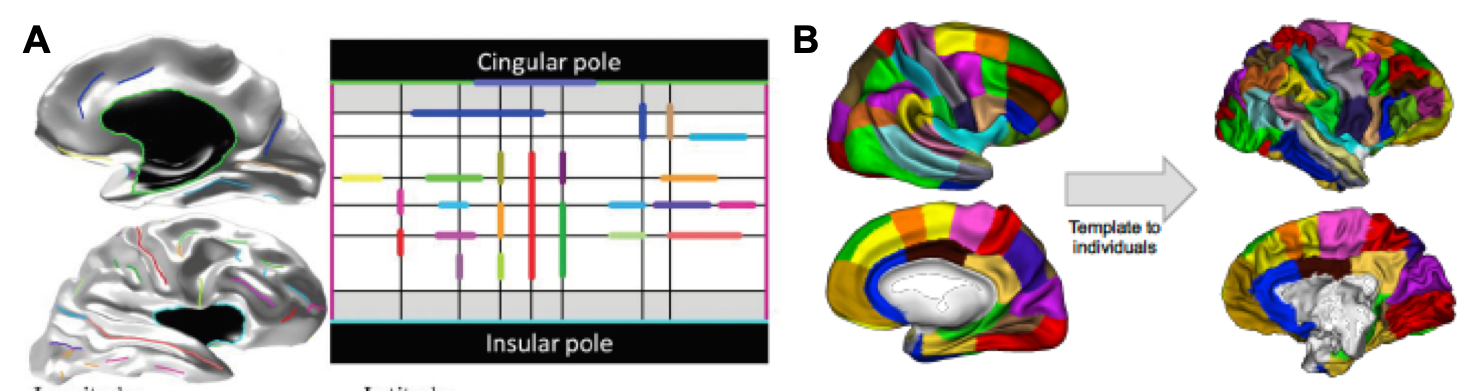
Based on the principle that homologous sets of cortical folds or sulci exist across human and non-human primate brains, a third potential approach involves the cross-species mapping of cortical surfaces based on common sulci that exist across species. Basically, this is accomplished by constructing an anatomical model of the cortex which describes the relative orientations, positions, and alignments of the various cortical sulci on the entire cortical surface (Auzias et al., 2013, IEEE Trans. On Medical Imaging). This model can be used to constrain a parameterisation of any individual’s cortical surface, and the resulting coordinate system can in turn be applied to compute correspondences between individual cortical surfaces within or across species. Here, the “common space” between species is essentially the relative layout of the various sulci on the cortical sheet, as captured by the parameterized cortical sulci model (Figure 10)
|
|---|
| Figure 10. A: Example of a human cortical surface and its sulcal lines (left) being projected on the cortical surface model (right). B: Coordinate-based mapping of a human parcellation (left, Auzias et al., 2016, Neuroimage) onto a chimpanzee brain (right) based on correspondences computed between the human (shown in A) and chimpanzee cortical model (not shown). |
Currently, the human cortical surface model has been constructed and implemented in Brainvisa. The template as well as tools and procedures for constructing human models can be found here. Ongoing work is performed to adapt the pipeline for constructing similar cortical models in non-human primate species (e.g. macaques, baboons and chimpanzees).
A. Why the interest in NHP neuroimaging?
B. What makes NHP MRI challenging?
C. Typical data analysis challenges
D. Structural data processing steps and PRIME-RE tools
E. Functional data processing steps and PRIME-RE tools
F. Diffusion data processing steps and PRIME-RE tools
G. Cross-species comparisons and PRIME-RE tools