Full documentation: here
Intro to AID-HS and installation videos: here
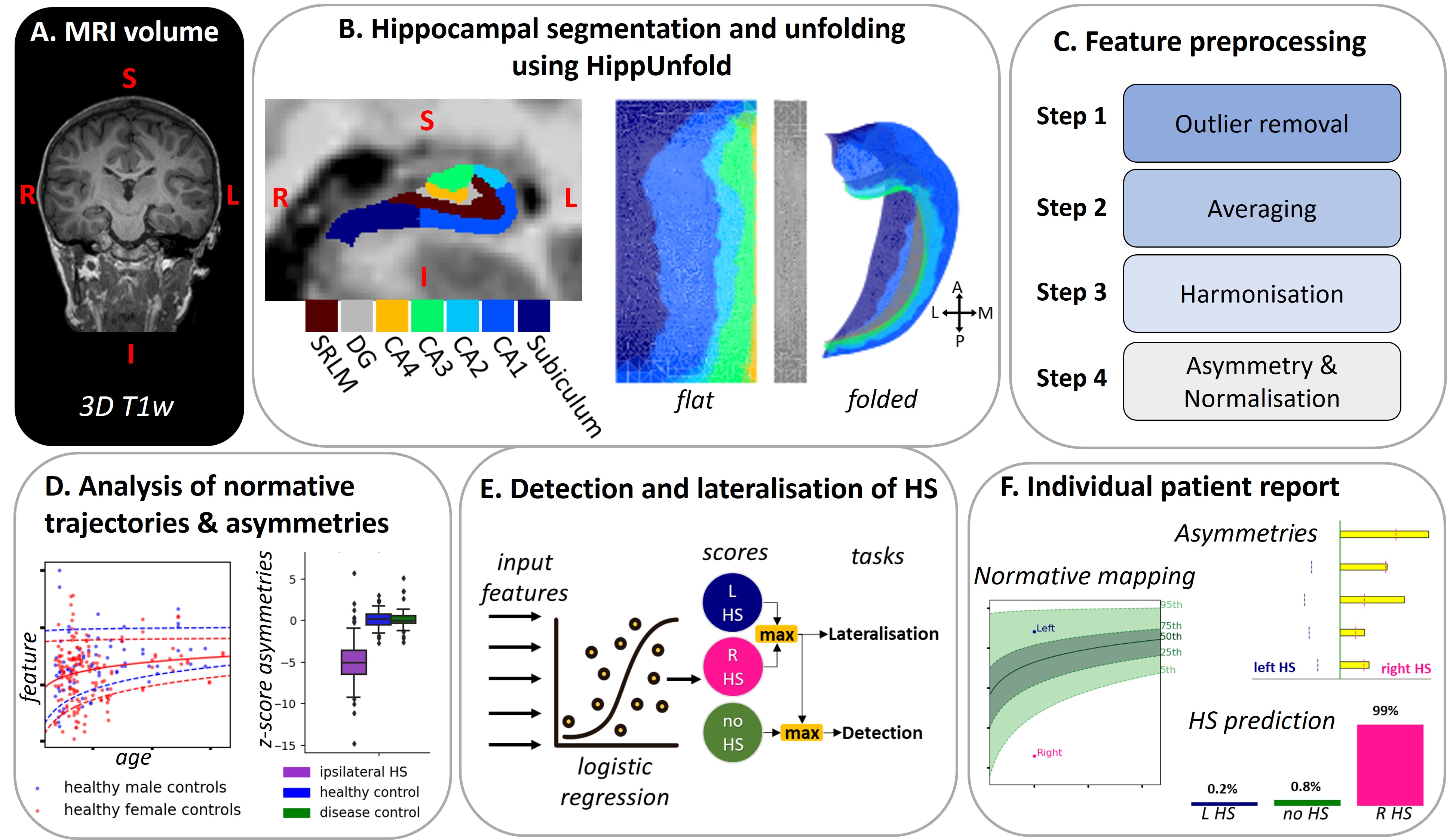
Automated and Interpretable Detection of Hippocampal Sclerosis
AID-HS extracts hippocampal volume- and surface-based features from T1w scans using HippUnfold, and provides an in-depth characterisation of hippocampal abnormalities as well as the automated detection and lateralisation of hippocampal sclerosis (HS). For more details please read our manuscript
Note:
- You will need the following demographic information (age at scan & sex) to run AID-HS on your patient's T1 MRI scan.
- AID-HS has been developed on T1w scans acquired at 3T. It has not yet been thoroughly evaluated on 1.5T and 7T data
SIGN UP TO THE AID-HS MAILING LIST:
We request that all AID-HS users sign up to the mailing list. If you are using AID-HS, please send an email to [email protected] with the subject 'Request to be added to the AID-HS mailing list' and provide use with your name and institute. This will ensure that we can update you about bug fixs and new releases.
EXISTING USERS: PLEASE UPDATE TO VERSION V1.0.1: We have released AID-HS V1.0.1 which fixes a couple of issues found by users. For more information about the release please see AID-HS V1.0.1. To update your code please follow the guidelines Updating AID-HS to V1.0.1 from our FAQ.
The AID-HS software is intended for research purposes only and has not been reviewed or approved by the Medicines and Healthcare products Regulatory Agency (MHRA), European Medicine Agency (EMA) or by any other agency. Any clinical application of the software is at the sole risk of the party engaged in such application. There is no warranty of any kind that the software will produce useful results in any way. Use of the software is at the recipient's own risk.
You can install and use the AID-HS pipeline with :
- docker container recommended for easy installation of the pipeline as all the prerequisite packages are already embeded into the container. Note: Dockers do not work on High Performance Computing (HCP) systems.
- singularity container - to run a container on High Performance Computing (HCP) systems.
- native installation: Not supported
YouTube tutorial available for the docker and singularity installation
FAQs If you have a question or if you are running into issues at any stage (installation/use/interpretation), have a look at our FAQs page as we may have already have a solution.
Once installed you will be able to use the AID-HS pipeline on your data following the steps:
- Prepare your data : guidelines
- (OPTIONAL) Compute the harmonisation parameters : guidelines
- Run the prediction pipeline: guidelines
- Interpret the results: guidelines
YouTube tutorials available to run the harmonisation step, to run the prediction pipeline and to interpret the pipeline results
What is the harmonisation process ?
Features extracted from MRI scans from different MRI scanners have systematic differences between them. To remove scanner related biases we recommend harmonising your MRI data to the MRI data that was used in the AID-HS manuscript. This harmonisation is required for each MRI scanner / T1 sequence you are using.
Notes:
- This step needs to be run only once, and requires data from at least 20 subjects acquired on the same scanner with the same T1 sequence and demographic information (e.g age and sex). See harmonisation instructions for more details.
- The AID-HS pipeline can also be run without harmonisation with no drop in performances. However, the characterisation of the hippocampal features compared to the normative growth curves will not be interpretable.
If you are using the AID-HS tool or part of the code, please cite:
An overview of the notebooks that we used to create the figures can be found here.
Mathilde Ripart, PhD \
Research Fellow at UCL Great Ormond Street Institute of Child Health
[email protected]