Process raw data from an Image Analysis GUI into biologically relevant features suitable for data mining and machine learning algorithms. Used to investigate the dynamic morphology of the cell segretation apparatus in saccharomyces cerevisiae.
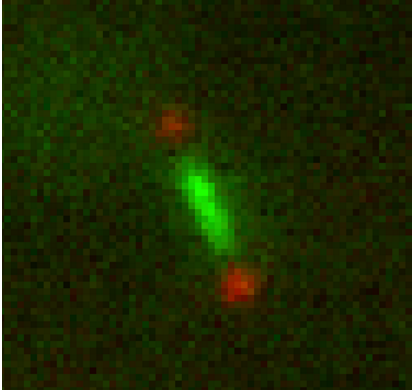
Features ideally used to identify phenotypes of four budding yeast mutants (SIR2∆, YKU80∆, BRN1-9 and wild-type) as they undergo mitosis on a dicentric plasmid.
This program takes tiff stacks of a cell undergoing mitosis with a LacO/GFP array on one half of a dicentric plasmid.
-
Track spindle pole bodies and plasmid signals using imageAnalyzer.m (it is a GUI) in the same directory as your tiff stacks.
-
Run loopDotMats.m in directory of one mutant. Example: sir2∆ = loopDotMats('sir2').
-
Concatenate all desired tables using vertcat().
From there, you can save the MATLAB table as a .csv, and from there the options for data analysis are endless. The world is your oyster.
See example images for raw images of data and example data from the analysis of these raw images.
Written in 2016 for the lab of Dr. Kerry Bloom, Chapel Hill, NC. (http://bloomlab.web.unc.edu/)
For relevant background information, check out this nifty paper http://bloomlab.web.unc.edu/files/2016/01/Thrower2001.pdf