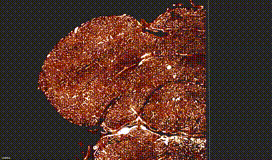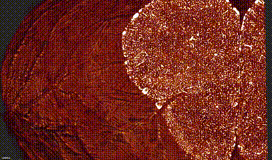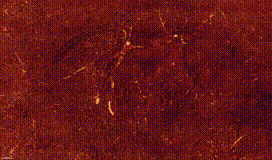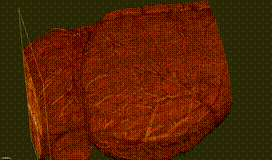
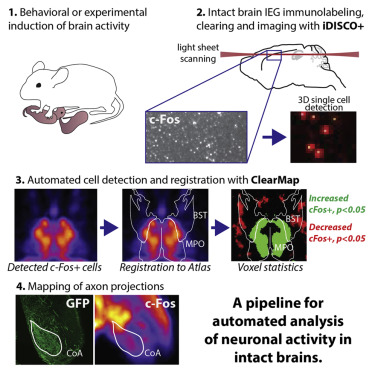
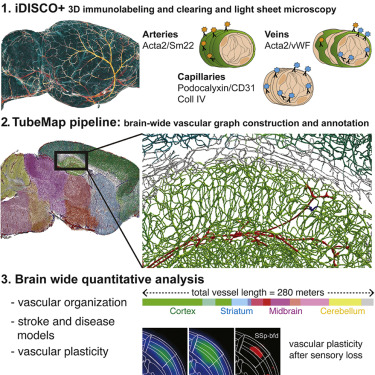
ClearMap is a toolbox for the analysis and registration of volumetric data from cleared tissues.
🆕 This is the new 2.1 version of ClearMap which includes a Graphical User Interface. To use it, make sure that the install_gui.sh script is executable and run it by opening a terminal in the ClearMap2 folder and typing ./run_gui.sh.
- Graphical user interface with many helper widgets
- Redesigned code with config based parameters to replace the scripts
- Atlas alignment improvements:
- Updated Allen atlas files (no more "No Label")
- Support for more atlases
- Support for hemispheres information
- Landmarks based registration
- Batch mode for processing or analysis
- New plots to visualise detected cells
- Various bug fixes
ClearMap's tool box includes
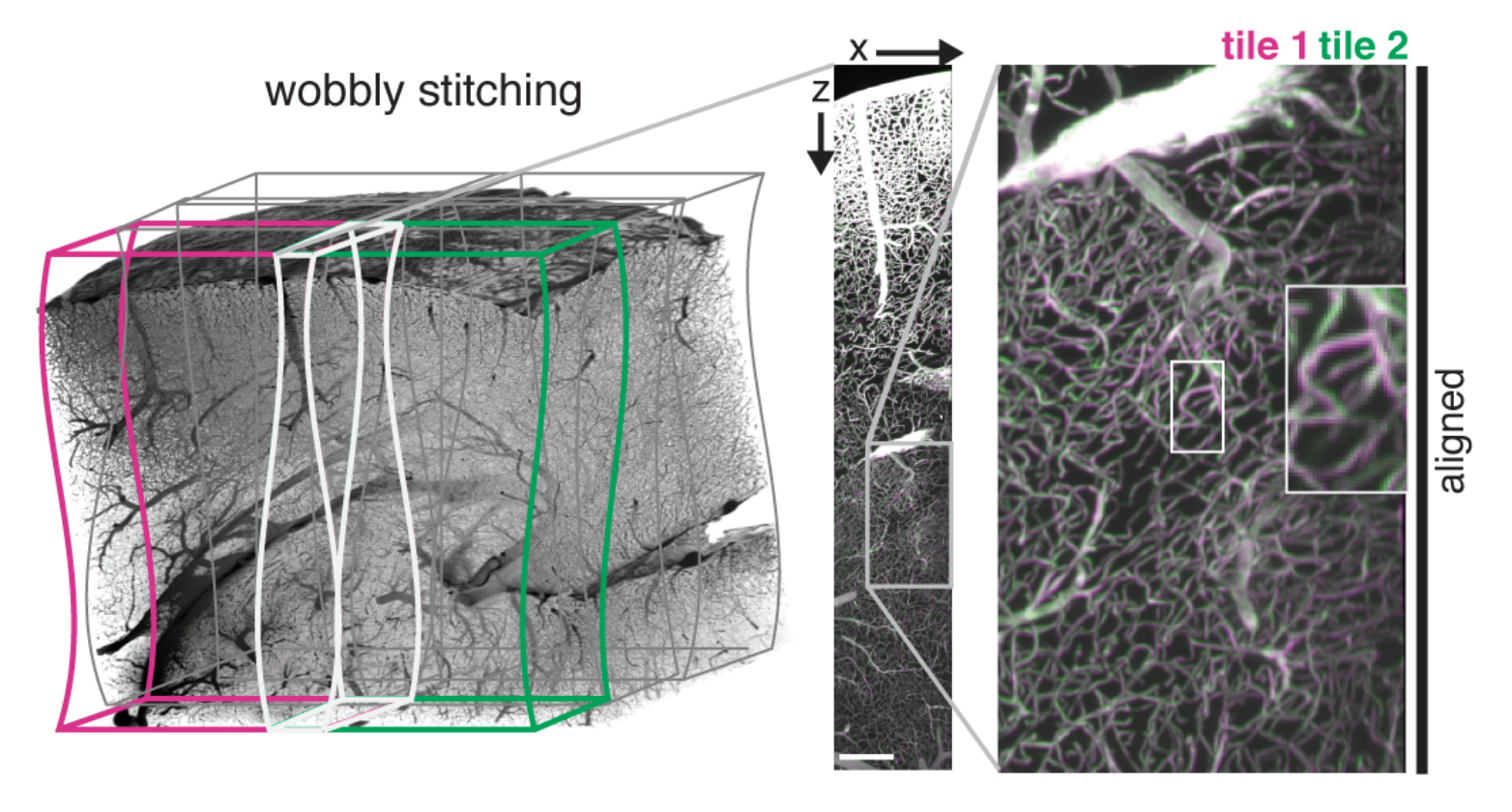
Stitch TB data sets non-rigidly.
Extract vasculature and other tubular networks from TB data.
Extract neuronal activity marker and cell shapes.
ClearMap has been designed to analyze O(TB) 3d datasets obtained via light sheet microscopy from iDISCO+ cleared tissue samples immunolabeled for proteins.
ClearMap has been written for mapping immediate early genes Renier et al. Cell 2016 as well as vasculature networks of whole mouse brains Kirst et al. Cell 2020
ClearMap tools may also be useful for data obtained with other types of microscopes, types of markers, clearing techniques, as well as other species, organs, or samples.
ClearMap is written in Python 3 and is designed to take advantage of parallel processing capabilities of modern workstations. We hope the open structure of the code will enable many new modules to be added to ClearMap to broaden the range of applications to different types of biological objects or structures.
Please refer to our documentation on how to install ClearMap.
Tutorials for TubeMap and CellMap are avaialbe as jupyer notebooks:
ClearMap comes with a full documentation.
For experimental protocols also refer to idisco.info
ClearMap has been featured in differnet articles, interviews and a TEDx talk:
See also our media gallery.
See who cites us:
and cite us if you use the sofware in any form:
@article{kirst2020mapping,
title={Mapping the fine-scale organization and plasticity of the brain vasculature},
author={Kirst, Christoph and Skriabine, Sophie and Vieites-Prado, Alba and Topilko, Thomas and Bertin, Paul and Gerschenfeld, Gaspard and Verny, Florine and Topilko, Piotr and Michalski, Nicolas and Tessier-Lavigne, Marc and others},
journal={Cell},
volume={180},
number={4},
pages={780--795},
year={2020},
publisher={Elsevier},
url={https://doi.org/10.1016/j.cell.2016.05.007}}
@article{renier2016mapping,
title={Mapping of brain activity by automated volume analysis of immediate early genes},
author={Renier, Nicolas and Adams, Eliza L and Kirst, Christoph and Wu, Zhuhao and Azevedo, Ricardo and Kohl, Johannes and Autry, Anita E and Kadiri, Lolahon and Venkataraju, Kannan Umadevi and Zhou, Yu and others},
journal={Cell},
volume={165},
number={7},
pages={1789--1802},
year={2016},
publisher={Elsevier},
url={https://doi.org/10.1016/j.cell.2020.01.028}}
ClearMap was originally designed and developed by Christoph Kirst.
Scripts and specific applications were developed by Nicolas Renier and Christoph Kirst.
Version 2.1 and GUI by Charly Rousseau and Etienne Doumazane with group analysis scripts contributed by Sophie Skriabine.
The deep vessel filling network was designed and created by Sophie Skriabine and integrated to ClearMap by Christoph Kirst.
The documentation was written by Christoph Kirst. and Nicolas Renier.
Contributions are very welcome.
This project is licensed under the GNU General Public License v3.0.
For other options contact the author Christoph Kirst ([email protected]).
Copyright © 2020 by Christoph Kirst
- Rewrite of the upper layers of software based on configuration files.
- Graphical user interface.
Rewrite of ClearMap 1.0 to handle larger data sets (TB). This version implements TubeMap
First version of ClearMap. Implements CellMap See https://github.com/ChristophKirst/ClearMap