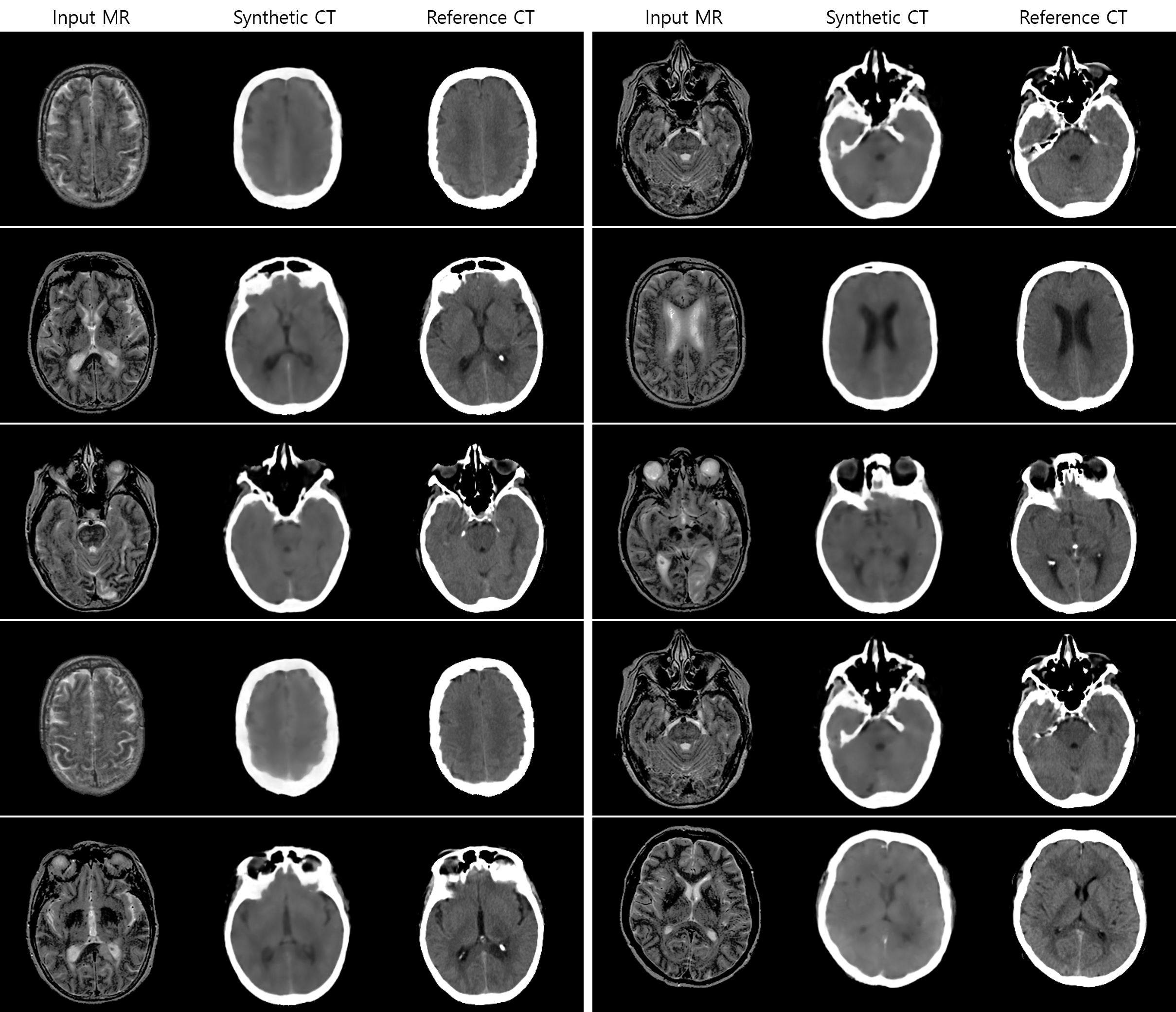
This repository is an implementation of "MR‐based synthetic CT generation using a deep convolutional neural network method." Medical physics 2017 by Xiao Han.
- tensorflow 1.13.1
- numpy 1.15.2
- opencv 4.1.0.25
- matplotlib 2.2.3
- pickleshare 0.7.4
- simpleitk 1.2.0
- scipy 1.1.0
- Six cross-validations
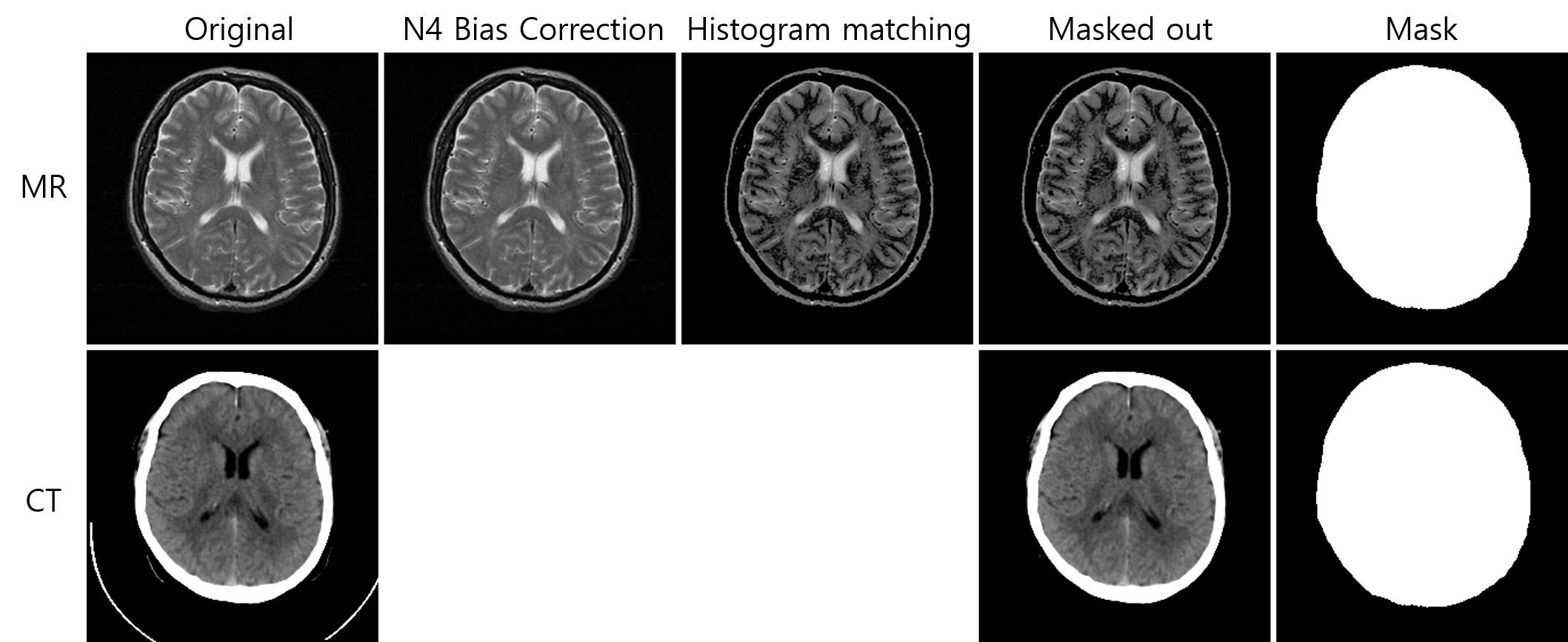
- Data preprocessing methods: N4 bias correction, histogram matching, and mask generation
- U-Net for CNN model
- Encoder of the U-Net is initialized by pretrained VGG16 weights
- Best model is saved based on the MAE evaluation in validation data
- Tensorboard visualization
- MAE, ME, MSE, and PCC metrics in test data
Download our toy dataset from here. This toy dataset just includes 367 paired images. We randomly divide data into training, validation, and test.
.
│ MRI-to-CT-DCNN-TensorFlow
│ ├── src
│ │ ├── dataset.py
│ │ ├── get_mask.py
│ │ ├── histogram_matching.py
│ │ ├── main.py
│ │ ├── model.py
│ │ ├── n4itk.py
│ │ ├── preprocessing.py
│ │ ├── solver.py
│ │ └── utils.py
│ Data
│ └── brain01
│ │ └── raw
│ Models_zoo
│ └── caffe_layers_value.pickle
Download the pretrained VGG16 weights from here (password:3ouy).
Use preprocessing.py to rectify N4 bias correction, histogram matching, and head mask generation. Example usage:
python preprocessing.py --delay=1 --is_save
data: dataset path for preprocessing, default:../../Data/brain01/rawtemp_id: template image id for histogram matching, default:2size: 'image width and height (width == height), default:256delay: interval time when showing image, default:1is_save: save processed image or not, default:False
- N4 bias field correction using N4ITK
- Histogram matching using Dynamic histogram warping algorithm
- Binary head mask using Otsu auto-thresholding
Use main.py to train a DCNN model. Example usage:
python main.py --is_train
gpu_index: gpu index if you have multiple gpus, default:0is_train: training or test mode, default:False (test mode)batch_size: batch size for one iteration, default:8dataset: dataset name, default:brain01learning_rate: learning rate, default:2e-4epoch: number of epochs, default:600print_freq: print frequency for loss information, default:100load_model: folder of saved model that you wish to continue training, (e.g. 20190411-2217), default:None
Use main.py to test the DCNN model. Example usage:
python main.py --load_model=folder/you/wish/to/test/e.g./20190411-2217
please refer to the above arguments.
Evaluation of the MAE, ME, MSE, and PCC in validation data during training process. Different color represents different model in six cross-validations.
Total loss, data loss and regularization term in each iteration.
MAE, ME, MSE, and PCC for six models and average performance.
@misc{chengbinjin2019DCNN,
author = {Cheng-Bin Jin},
title = {MRI-to-CT-DCNN-Tensorflow},
year = {2019},
howpublished = {\url{https://github.com/ChengBinJin/MRI-to-CT-DCNN-TensorFlow},
note = {commit xxxxxxx}
}
- Han, Xiao. "MR‐based synthetic CT generation using a deep convolutional neural network method." Medical physics 44.4 (2017): 1408-1419.
- Tustison, Nicholas J., et al. "N4ITK: improved N3 bias correction." IEEE transactions on medical imaging 29.6 (2010): 1310.
- Cox, Ingemar J., Sébastien Roy, and Sunita L. Hingorani. "Dynamic histogram warping of image pairs for constant image brightness." Proceedings., International Conference on Image Processing. Vol. 2. IEEE, 1995.
- Otsu, Nobuyuki. "A threshold selection method from gray-level histograms." IEEE transactions on systems, man, and cybernetics 9.1 (1979): 62-66.
Copyright (c) 2018 Cheng-Bin Jin. Contact me for commercial use (or rather any use that is not academic research) (email: [email protected]). Free for research use, as long as proper attribution is given and this copyright notice is retained.