- NEW Releases
- TOP contributors
- About Us
- Our model
- Pipelines
- Tools
ccbrpipelinermodule release history on BIOWULF- Latest Releases of pipelines/tools:
- Citation
| Repo Name | Release Name | Release Date | Open Issues |
|---|---|---|---|
| Dockers2 | v0.1.0 | 2024-11-12 | 3 |
| actions | v0.2.1 | 2024-11-11 | 5 |
| XAVIER | v3.1.2 | 2024-11-05 | 8 |
| RENEE | v2.6.1 | 2024-10-25 | 16 |
| parkit | v2.1.0 | 2024-10-16 | 2 |
| CRISPIN | v1.0.1 | 2024-10-16 | 15 |
| CARLISLE | v2.6.1 | 2024-10-14 | 12 |
| ESCAPE | v1.1.4 | 2024-09-23 | 0 |
| CHARLIE | v0.11.0 | 2024-09-16 | 15 |
| ASPEN | v1.0.2 | 2024-09-12 | 4 |
| CHAMPAGNE | v0.4.0 | 2024-09-11 | 30 |
| Tools | v0.1.1 | 2024-09-10 | 9 |
| reports | v0.2.2 | 2024-08-22 | 9 |
| User | Total Commits | Commits in Last Month | Commits in Last 6 Months |
|---|---|---|---|
| kelly-sovacool | 4750 | 322 | 1535 |
| kopardev | 4444 | 186 | 393 |
| slsevilla | 1700 | 0 | 0 |
| skchronicles | 1074 | 0 | 0 |
| dnousome | 666 | 4 | 99 |
| kcgfarb | 629 | 63 | 212 |
| samarth8392 | 460 | 8 | 152 |
| finneyr | 348 | 0 | 8 |
| jlac | 307 | 0 | 0 |
| kvaldez | 222 | 0 | 0 |
- 👋 Hi, we're the @CCBR, a group of bioinformatics analysts and engineers
- 📖 We build flexible, reproducible, workflows for next-generation sequencing data
- 💡 We collaborate with CCR PIs
- 📫 You can reach us at [email protected]
- 🏁 Check out our release history
- 🔗 Our Zenodo community
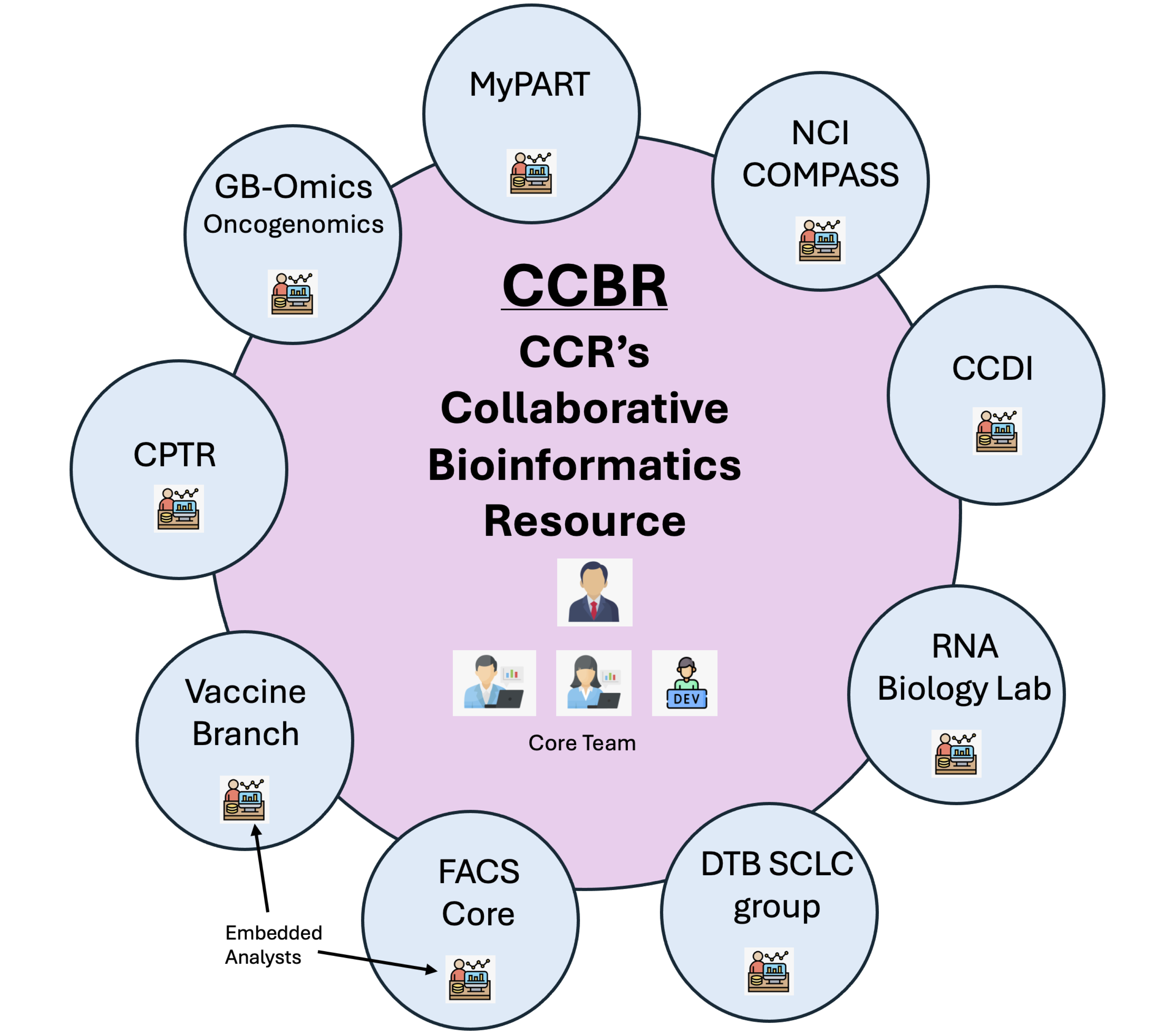
The CCR Collaborative Bioinformatics Resource (CCBR) operates as a crucial support system within the National Cancer Institute, particularly for researchers at the Center for Cancer Research (CCR). CCBR brings together a team of bioinformatics experts from various institutions, including the CCR Office of Science and Technology Resources (OSTR), Frederick National Laboratory for Cancer Research (FNLCR), and the Center for Biomedical Informatics and Information Technology (CBIIT). This collaborative model allows CCBR to offer a wide array of bioinformatics services, ranging from data analysis and interpretation to software development and training.
CCBR's working model emphasizes accessibility and collaboration. Researchers at CCR can easily request bioinformatics support through a formal project submission process. This process is designed to streamline access to bioinformatics resources, ensuring that projects receive timely and appropriate expertise. The CCBR team offers personalized consultations, both in-person and virtual, to understand the specific needs of each research project, facilitating a tailored approach that enhances the quality and impact of scientific research within the cancer research community.
The CCBR comprises of the core team and the embedded team. These teams serve different, but similar roles within the CCBR's support structure.
-
The core team consists of bioinformatics experts who provide centralized support to researchers, offering services such as data analysis, software development, and training across a broad range of projects. They are available to assist any CCR researcher who submits a request for bioinformatics help.
-
The embedded team members are bioinformaticians who are integrated within specific research groups or labs. Their role is to provide more focused, day-to-day bioinformatics support tailored to the specific needs of the research group they are embedded in. This allows for a deeper collaboration and a more nuanced understanding of the ongoing research projects within that group.
For details, please visit CCBR's official website.
CCBR offers end-to-end analysis pipelines for NGS data analysis.
RHEL8 BIOWULF updates:
In late 2023, BIOWULF migrated to a new operating system, RHEL8. This migration rendered the Legacy functionality of CCBRPipeliner un-usable. We have been (and will continue to) work dilegently to bring
ccbrpipelinersuite of pipelines back on-line for our Biowulf users. At the same time, we are also taking this opportunity to not only increase our repetoire of pipelines but also modernize and containerize our end-to-end analysis offerings. These changes will minimize, if not eliminate, the pipelines' dependencies on other Biowulf modules and makeccbrpipeliner"operating system and HPC" - agnostic, thereby making it shareable with collaborators, and runnable on other HPCs (like FRCE) and beyond.
Here is a list of our prominent pipelines and their release schedule on BIOWULF:
| Data Type | Pipeline Name | CLI* availability date | GUI* availability date |
|---|---|---|---|
| RNASeq1 | RENEE |
July 3rd 2023 | July 14th 2023 |
| WESSeq2 | XAVIER |
July 21th 2023 | Sep 1st 2023 |
| ATACSeq3 | ASPEN |
November 30th 2023 | TBD |
| ChIPSeq4 | CHAMPAGNE |
October 15th 2023 | TBD |
| CRISPRSeq5 | CRISPIN |
September 31st 2023 | TBD |
| CUT&RunSeq6 | CARLISLE |
October 31st 2023 | TBD |
| EV-Seq10 | ESCAPE |
March 26th, 2024 | TBD |
| circRNASeq7 | CHARLIE |
Jul 31st 2024 | TBD |
| scRNASeq8 | SINCLAIR |
Sep 30th 2024 | TBD |
| WGSSeq9 | LOGAN |
Sep 30th 2024 | TBD |
| spatialSeq11 | SPENCER |
TBD | TBD |
* CLI = Command Line Interface * GUI = Graphical User Interface
1 RENEE=Rna sEquencing aNalysis pipElinE starts with raw fastq files and ends with counts matrix. Downstream DEG support will be added at a later date. In the mean time you can use NIDAP or iDEP for DEG analysis.
2 XAVIER=eXome Analysis and Variant explorER will be soon available on Biowulf.
3 ASPEN=Atac Seq PipEliNe has limited support for differential ATACSeq signal analysis. CCBR has other pipelines for footprinting analysis like TOBIAS. Please reach out for details.
4 CHAMPAGNE=CHromAtin iMmuno PrecipitAtion sequencinG aNalysis pipEline. CCBR plans to completely revamp ChIPSeq and may not be available until Q4 of 2023. In the interim, we recommend using the ENCODE pipeline on biowulf for ChIPSeq analsyis.
5 CRISPIN=CRISPr screen sequencing analysis pipelINe( previously called, CRUISE=Crispr scReen seqUencIng analySis pipEline ). CRISPRSeq analysis with MAGeCK, drugZ and BAGEL2.
6 CARLISLE=Cut And Run anaLysIS pipeLinE supports human and mouse samples with (recommended) or without spike-ins.
7 CHARLIE=Circrnas in Host And viRuses anaLysis pIpEline finds known and novel circRNAs in human/mouse + virus genomes. Differential circRNA analysis is planned for future.
8 SINCLAIR=SINgle CelL AnalysIs Resource addresses various single cell modalities... eg. single-cell expression, CITESeq, TCR-Seq, etc.
9 LOGAN=whoLe genOme-sequencinG Analysis pipeliNe will soon be CCBR's newest offering.
10 ESCAPE=Extracellular veSiCles rnAseq PipelinE.
11 SPENCER=SPatial seqeENCing Resource.
For any other datatype or pipeline, please email 📫 us directly to get the conversation started!
In additions to end-to-end analysis pipelines, the CCBR dev team also builds tools for data management, meta-data management, APIs, user management, etc. Here are some examples:
module load ccbrpipeliner loads default release of ccbrpipeliner. Each release comprises of a unique combination of the version numbers of the different pipelines offered as part of the ccbrpipeliner suite.
| Release | Tool versions | Released on | Decommissioned on |
|---|---|---|---|
| 1 | RENEE v2.1 @# | July, 10th 2023 | July, 14th 2023 |
| 2 | RENEE v2.2 @# | July, 14th 2023 | September, 5th 2023 |
| 3 | RENEE v2.2 @#, XAVIER v2.0 @ | July, 21st 2023 | - |
| 4 | RENEE v2.5 @#, XAVIER v3.0 @# | September, 5th 2023 | - |
| 5 | RENEE v2.5 @#, XAVIER v3.0 @#, CARLISLE v2.4 @, CHAMPAGNE v0.2 @, CRUISE v0.1 @, spacesavers2 v0.10 @, permfix v0.6 @ | October, 27th 2023 | - |
| 6* | RENEE v2.5 @#, XAVIER v3.0 @#, CARLISLE v2.4 @, CHAMPAGNE v0.3 @, CRUISE v0.1 @, ASPEN v1.0 @, spacesavers2 v0.12 @, permfix v0.6 @ | February, 29th 2024 | - |
* = Current DEFAULT version on BIOWULF
@ = CLI available
# = GUI available
module load ccbrpipelineris also available on HELIX. It only loads the tools and not the pipelines as HELIX does not have a job scheduler
| Repo Name | Release Name | Release Date | Open Issues |
|---|---|---|---|
| Dockers2 | v0.1.0 | 2024-11-12 | 3 |
| actions | v0.2.1 | 2024-11-11 | 5 |
| XAVIER | v3.1.2 | 2024-11-05 | 8 |
| RENEE | v2.6.1 | 2024-10-25 | 16 |
| parkit | v2.1.0 | 2024-10-16 | 2 |
| CRISPIN | v1.0.1 | 2024-10-16 | 15 |
| CARLISLE | v2.6.1 | 2024-10-14 | 12 |
| ESCAPE | v1.1.4 | 2024-09-23 | 0 |
| CHARLIE | v0.11.0 | 2024-09-16 | 15 |
| ASPEN | v1.0.2 | 2024-09-12 | 4 |
| CHAMPAGNE | v0.4.0 | 2024-09-11 | 30 |
| Tools | v0.1.1 | 2024-09-10 | 9 |
| reports | v0.2.2 | 2024-08-22 | 9 |
| spacesavers2 | v0.14.0 | 2024-07-16 | 4 |
| permfix | v0.6.4 | 2024-05-07 | 0 |
| journal-club | v0.1.0 | 2024-05-07 | 2 |
| CCBR_tobias | v0.3.0 | 2024-04-12 | 3 |
| METRO | v2.1 | 2024-03-28 | 2 |
| CCBR-1144 | v1.0.0 | 2024-03-04 | 0 |
| TRANQUIL | v0.2.1 | 2024-02-22 | 1 |
| ccbr1271_ERVpipeline | v1.0.3 | 2024-02-21 | 1 |
| nf-modules | v0.1.0 | 2023-11-29 | 11 |
| SINCLAIR | v0.2.0 | 2023-11-01 | 30 |
| CRISPRAnnotation | v1.0 | 2023-10-19 | 0 |
| SharanLab | v1.0.0 | 2023-07-18 | 0 |
| Pipeliner | v4.0.7 | 2023-05-09 | 24 |
| MAPLE | v1.0.1 | 2023-02-27 | 0 |
| DTB_ExomeSeq | v1.0 | 2022-06-22 | 0 |
| ASCENT | v0.1.1 | 2022-01-04 | 2 |
| Antitumor-activity-of-entinostat-plus-NHS-IL12 | v.1.0 | 2021-06-01 | 0 |
| CCBR_circRNA_AmpliconSeq | v0.1.1 | 2021-03-24 | 0 |
| rNA | v1.0.0 | 2021-01-21 | 0 |
| l2p | v0.0.3 | 2020-07-13 | 0 |
| MAAPster | v2.0.0 | 2020-04-27 | 0 |
| ChIP-Seq-Pipeline | alpha2 | 2016-10-01 | 2 |
Most of our end-to-end pipelines which have been used in published research work have been made available to the entire bioinformatics community via a Zenodo DOI. Please feel free to visit our Zenodo community page. And if you use our pipelines, don't forget to cite us!