-
Notifications
You must be signed in to change notification settings - Fork 2
Installation
iBrowser can be downloaded as an self contained (Virtual Box) virtual machine using BitTorrent Sync and this read-only key: key (860Mb).
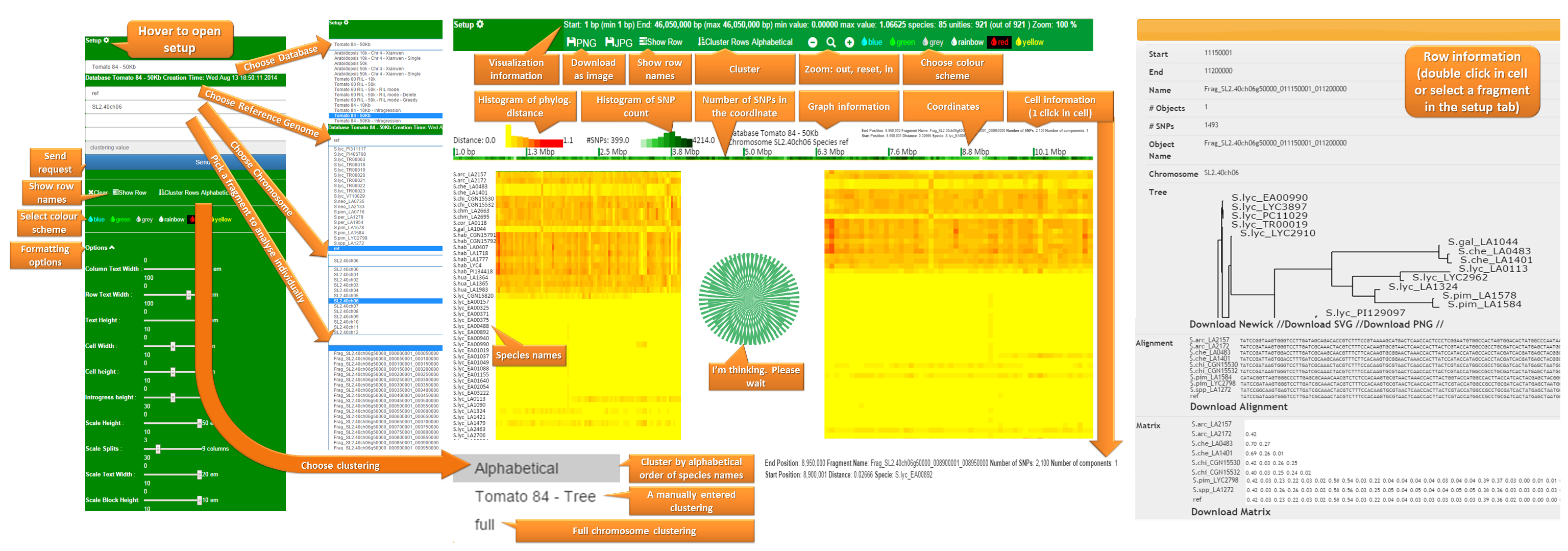
A manual can be found here
The data used in the iBrowser paper can be downloaded using BitTorrent Sync using the following read-only key: key.
The data consists of:
total 39G
9.1G arabidopsis_50k.sqlite
1.5K arabidopsis_50k.sqlite.nfo
6.4G RIL_10k.sqlite
1.5K RIL_10k.sqlite.nfo
1.5K RIL_50k_mode_ril_delete_greedy.sqlite.nfo
1.6G RIL_50k_mode_ril_delete.sqlite
1.5K RIL_50k_mode_ril_delete.sqlite.nfo
995M RIL_50k_mode_ril_greedy.sqlite
1.5K RIL_50k_mode_ril_greedy.sqlite.nfo
1.6G RIL_50k_mode_ril.sqlite
1.5K RIL_50k_mode_ril.sqlite.nfo
1.6G RIL_50k.sqlite
1.5K RIL_50k.sqlite.nfo
1.5K RIL.customorder
67M tom84_10k_introgression.sqlite
1.5K tom84_10k_introgression.sqlite.nfo
11G tom84_10k.sqlite
1.5K tom84_10k.sqlite.nfo
27M tom84_50k_introgression.sqlite
1.5K tom84_50k_introgression.sqlite.nfo
3.1G tom84_50k.sqlite
1.5K tom84_50k.sqlite.nfo
3.0K tom84.customorder
3.9G tom84_genes.sqlite
1.5K tom84_genes.sqlite.nfoThe Web User Interface (Web UI) works in Chrome 37+ and Firefox 33+. The UI behaviour in iExplorer is erratic and therefore we discourage the use of this web browser. We haven't tested Opera and Safari web browsers, feedback is welcome.
for standalone:
docker run -it --rm \
-v $PWD/data:/var/www/ibrowser/data \
-p 127.0.0.1:10000:10000 \
--name ibrowser \
sauloal/introgressionbrowser ./ibrowser.py data/for local copy,run:
git clone [email protected]:sauloal/introgressionbrowser.git
cd introgressionbrowser
docker run -it --rm \
-v $PWD:/var/www/ibrowser \
-v $PWD/data:/var/www/ibrowser/data \
-p 127.0.0.1:10000:10000 \
--name ibrowser \
sauloal/introgressionbrowser_local ./ibrowser.py data/Open your browser at 127.0.0.1:10000
replace 127.0.0.1 for 0.0.0.0 in the command line if you want others in your network to be able to access your iBrowser instance.
replace the -it for -d to run in the background
The virtual machine should run automatically. The only procedure is to share your data folder (in your host computer) as "DATA". A step-by-step manual can be found here. In case you want/need to do it manually, please find bellow instructions:
wget http://download.virtualbox.org/virtualbox/4.3.6/VBoxGuestAdditions_4.3.6.iso
mkdir vbox
mount VBoxGuestAdditions_4.3.6.iso vbox
cd vbox
./VBoxLinuxAdditions.run
cd ..
umount vbox
edit /etc/fstab adding
data /media/data vboxsf re 0 0
mount -a
ls /media/datacurrently there's a bug in vmware which doesn't allows for the mounting of shared folders. For this reason vmware is not currently supported
mkdir /mnt/cdrom
mount /dev/cdrom /mnt/cdrom
mkdir ~/vm
cd ~/vm
tar xvf /dev/cdrom/VMwareTools-9.6.1-1378637.tar.gz
cd vmware-tools-distrib
./vmware-install.pl -d
cd ../..
rm -rf vm
ls /mnt/hgfs/dataClone or download Introgression Browser.
git clone https://github.com/sauloal/introgressionbrowserapt-get install -y -f build-essential checkinstall openssl sqlite3 libsqlite3-dev \
libfreetype6 libfreetype6-dev zlib1g-dev libjpeg62 libjpeg62-dev \
pkg-config libblas-dev liblapack-dev gfortran zlib1g-dev
apt-get install -y -f python-setuptools python-dev python-numpy python-scipy \
python-matplotlib python-pandas python-sympy python-pip python-imaging \
python-numpypip install --requirement requirements.txtOR
easy_install --user flask
easy_install --user ete2
easy_install --user sqlalchemy
easy_install --user Flask-SQLAlchemy
easy_install --user pysha3
easy_install --user pycryptoIf not possible to install python libraries system wide with apt-get, install also:
easy_install --user Pillow
easy_install --user Image
easy_install --user numpy
easy_install --user scipy
easy_install --user matplotlib
easy_install --user MySQL-pythonBesides all standalone dependencies.
apt-get install -y -f libapache2-mod-wsgi apache2 - Besides all standalone dependencies. *
Install pypy (Optional, but speeds up analysis)
This project is maintained by Saulo Aflitos ( GitHub and LinkedIn ) with support from Applied Bioinformatics and WageningenUR
<script type="text/javascript">var gaJsHost = (("https:" == document.location.protocol) ? "https://ssl." : "http://www."); document.write(unescape("%3Cscript src='" + gaJsHost + "google-analytics.com/ga.js' type='text/javascript'%3E%3C/script%3E"));</script> <script type="text/javascript">try { var pageTracker = _gat._getTracker("UA-5291039-9"); pageTracker._trackPageview(); } catch(err) {}</script>--Get Data
---Installation
----Docker
-----Virtual Machine
------VirtualBox
------VMWare
-----Manually
------Getting the code
------Global dependencies
-------Visualization
-------Standalone
--------Install Linux dependencies
--------Install Python dependencies
-------Apache
--------Install Apache dependencies
-------Calculations
-Running
--Running Visualization Server
--Running Calculations
---General
---Input Data
---Run
----Automatically
-----Examples
----Manually
-----Merging
-----Splitting
-----Cleaning
-----Phylogeny
-----Extraction
-----Database creation