-
Notifications
You must be signed in to change notification settings - Fork 7
/
napari-aicsimageio.json
410 lines (410 loc) · 15.3 KB
/
napari-aicsimageio.json
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
{
"name": "napari-aicsimageio",
"display_name": "napari-aicsimageio",
"visibility": "public",
"icon": "",
"categories": [],
"schema_version": "0.1.0",
"on_activate": null,
"on_deactivate": null,
"contributions": {
"commands": [
{
"id": "napari-aicsimageio.get_reader",
"title": "Get AICSImageIO Reader",
"python_name": "napari_aicsimageio.core:get_reader",
"short_title": null,
"category": null,
"icon": null,
"enablement": null
}
],
"readers": [
{
"command": "napari-aicsimageio.get_reader",
"filename_patterns": [
"*.1sc",
"*.2fl",
"*.3fr",
"*.acff",
"*.acqp",
"*.afi",
"*.afm",
"*.aim",
"*.al3d",
"*.ali",
"*.am",
"*.amiramesh",
"*.ano",
"*.apl",
"*.arf",
"*.array-like",
"*.arw",
"*.avi",
"*.bay",
"*.bif",
"*.bin",
"*.bip",
"*.bmp",
"*.bmq",
"*.bsdf",
"*.bufr",
"*.bw",
"*.c01",
"*.cap",
"*.cat",
"*.cfg",
"*.ch5",
"*.cif",
"*.cine",
"*.cr2",
"*.crw",
"*.cs1",
"*.csv",
"*.ct",
"*.ct.img",
"*.cur",
"*.cut",
"*.cxd",
"*.czi",
"*.dat",
"*.db",
"*.dc2",
"*.dcm",
"*.dcr",
"*.dcx",
"*.dds",
"*.df3",
"*.dicom",
"*.dm2",
"*.dm3",
"*.dng",
"*.drf",
"*.dsc",
"*.dti",
"*.dv",
"*.ecw",
"*.emf",
"*.eps",
"*.epsi",
"*.erf",
"*.exp",
"*.exr",
"*.fake",
"*.fdf",
"*.fff",
"*.ffr",
"*.fid",
"*.fit",
"*.fits",
"*.flc",
"*.flex",
"*.fli",
"*.fpx",
"*.frm",
"*.ftc",
"*.fts",
"*.ftu",
"*.fz",
"*.g3",
"*.gbr",
"*.gdcm",
"*.gel",
"*.gif",
"*.gipl",
"*.grey",
"*.grib",
"*.h5",
"*.hdf",
"*.hdf5",
"*.hdp",
"*.hdr",
"*.hed",
"*.his",
"*.htd",
"*.htm",
"*.html",
"*.hx",
"*.i2i",
"*.ia",
"*.icns",
"*.ico",
"*.ics",
"*.ids",
"*.iff",
"*.iim",
"*.iiq",
"*.im",
"*.im3",
"*.img",
"*.imggz",
"*.ims",
"*.inf",
"*.inr",
"*.ipl",
"*.ipm",
"*.ipw",
"*.j2c",
"*.j2k",
"*.jfif",
"*.jif",
"*.jng",
"*.jp2",
"*.jpc",
"*.jpe",
"*.jpeg",
"*.jpf",
"*.jpg",
"*.jpk",
"*.jpx",
"*.jxr",
"*.k25",
"*.kc2",
"*.kdc",
"*.klb",
"*.koa",
"*.l2d",
"*.labels",
"*.lbm",
"*.lei",
"*.lfp",
"*.lfr",
"*.lif",
"*.liff",
"*.lim",
"*.lms",
"*.lsm",
"*.mdb",
"*.mdc",
"*.mef",
"*.mgh",
"*.mha",
"*.mhd",
"*.mic",
"*.mkv",
"*.mnc",
"*.mnc2",
"*.mng",
"*.mod",
"*.mos",
"*.mov",
"*.mp4",
"*.mpeg",
"*.mpg",
"*.mpo",
"*.mrc",
"*.mri",
"*.mrw",
"*.msp",
"*.msr",
"*.mtb",
"*.mvd2",
"*.naf",
"*.nd",
"*.nd2",
"*.ndpi",
"*.ndpis",
"*.nef",
"*.nhdr",
"*.nia",
"*.nii",
"*.nii.gz",
"*.niigz",
"*.npz",
"*.nrrd",
"*.nrw",
"*.obf",
"*.oib",
"*.oif",
"*.oir",
"*.ome",
"*.ome.tif",
"*.ome.tiff",
"*.orf",
"*.par",
"*.pbm",
"*.pcd",
"*.pcoraw",
"*.pct",
"*.pcx",
"*.pef",
"*.pfm",
"*.pgm",
"*.pic",
"*.pict",
"*.png",
"*.pnl",
"*.ppm",
"*.pr3",
"*.ps",
"*.psd",
"*.ptx",
"*.pxn",
"*.pxr",
"*.qptiff",
"*.qtk",
"*.r3d",
"*.raf",
"*.ras",
"*.raw",
"*.rcpnl",
"*.rdc",
"*.rec",
"*.rgb",
"*.rgba",
"*.rw2",
"*.rwl",
"*.rwz",
"*.scan",
"*.scn",
"*.sdt",
"*.seq",
"*.sif",
"*.sld",
"*.sm2",
"*.sm3",
"*.spc",
"*.spe",
"*.spi",
"*.sr2",
"*.srf",
"*.srw",
"*.st",
"*.sti",
"*.stk",
"*.stp",
"*.svs",
"*.swf",
"*.sxm",
"*.targa",
"*.tfr",
"*.tga",
"*.thm",
"*.tif",
"*.tiff",
"*.tim",
"*.tnb",
"*.top",
"*.txt",
"*.v",
"*.vff",
"*.vms",
"*.vsi",
"*.vtk",
"*.vws",
"*.wap",
"*.wat",
"*.wav",
"*.wbm",
"*.wbmp",
"*.wdp",
"*.webp",
"*.wlz",
"*.wmf",
"*.wmv",
"*.wpi",
"*.xbm",
"*.xdce",
"*.xml",
"*.xpm",
"*.xqd",
"*.xqf",
"*.xv",
"*.xys",
"*.zfp",
"*.zfr",
"*.zip",
"*.zpo",
"*.zvi"
],
"accepts_directories": false
}
],
"writers": null,
"widgets": null,
"sample_data": null,
"themes": null,
"menus": {},
"submenus": null,
"keybindings": null,
"configuration": []
},
"package_metadata": {
"metadata_version": "2.1",
"name": "napari-aicsimageio",
"version": "0.7.2",
"dynamic": null,
"platform": null,
"supported_platform": null,
"summary": "AICSImageIO bindings for napari",
"description": "# napari-aicsimageio\n\n[](https://github.com/AllenCellModeling/napari-aicsimageio/raw/main/LICENSE)\n[](https://github.com/AllenCellModeling/napari-aicsimageio/actions)\n[](https://codecov.io/gh/AllenCellModeling/napari-aicsimageio)\n\nAICSImageIO bindings for napari\n\n---\n\n## Features\n\n- Supports reading metadata and imaging data for:\n - `OME-TIFF`\n - `TIFF`\n - `CZI` (Zeiss)\n - `LIF` (Leica)\n - `ND2` (Nikon)\n - `DV` (DeltaVision)\n - Any formats supported by [aicsimageio](https://github.com/AllenCellModeling/aicsimageio)\n - Any formats supported by [bioformats](https://github.com/tlambert03/bioformats_jar)\n - `SLD` (Slidebook)\n - `SVS` (Aperio)\n - [Full List](https://docs.openmicroscopy.org/bio-formats/6.5.1/supported-formats.html)\n - Any additional format supported by [imageio](https://github.com/imageio/imageio)\n - `PNG`\n - `JPG`\n - `GIF`\n - `AVI`\n - [Full List](https://imageio.readthedocs.io/en/v2.4.1/formats.html)\n\n_While upstream `aicsimageio` is released under BSD-3 license, this plugin is released under GPLv3 license because it installs all format reader dependencies._\n\n## Installation\n\n**Stable Release:** `pip install napari-aicsimageio` or `conda install napari-aicsimageio -c conda-forge`<br>\n**Development Head:** `pip install git+https://github.com/AllenCellModeling/napari-aicsimageio.git`\n\n### Reading Mode Threshold\n\nThis image reading plugin will load the provided image directly into memory if it meets\nthe following two conditions:\n\n1. The filesize is less than 4GB.\n2. The filesize is less than 30% of machine memory available.\n\nIf either of these conditions isn't met, the image is loaded in chunks only as needed.\n\n### Use napari-aicsimageio as the Reader for All File Formats\n\nIf you want to force napari to always use this plugin as the reader for all file formats,\ntry running this snippet:\n\n```python\nfrom napari.settings import get_settings\n\nget_settings().plugins.extension2reader = {'*': 'napari-aicsimageio', **get_settings().plugins.extension2reader}\n```\n\nFor more details, see [#37](https://github.com/AllenCellModeling/napari-aicsimageio/issues/37).\n\n## Examples of Features\n\n#### General Image Reading\n\nAll image file formats supported by\n[aicsimageio](https://github.com/AllenCellModeling/aicsimageio) will be read and all\nraw data will be available in the napari viewer.\n\nIn addition, when reading an OME-TIFF, you can view all OME metadata directly in the\nnapari viewer thanks to `ome-types`.\n\n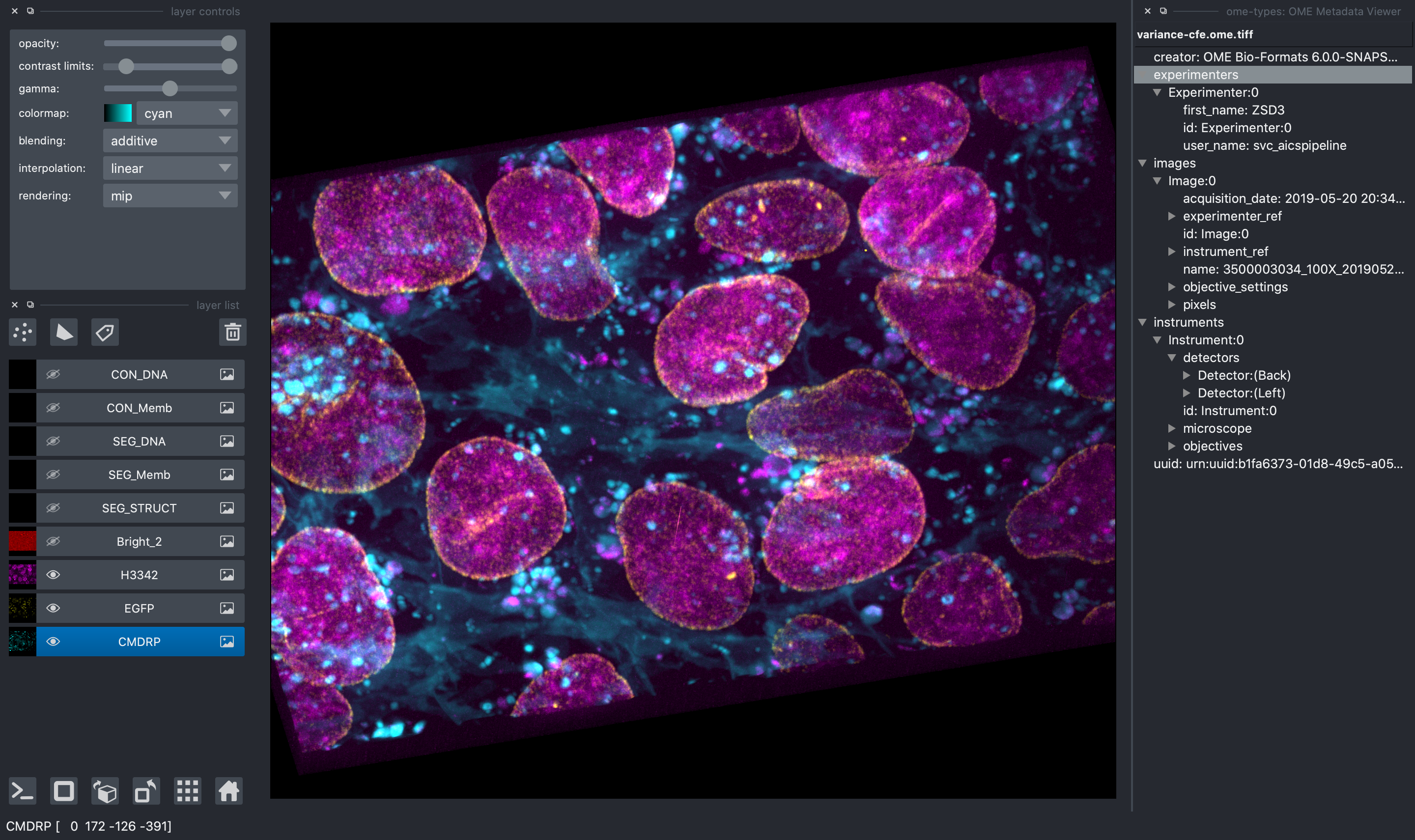\n\n#### Multi-Scene Selection\n\nWhen reading a multi-scene file, a widget will be added to the napari viewer to manage\nscene selection (clearing the viewer each time you change scene or adding the\nscene content to the viewer) and a list of all scenes in the file.\n\n\n\n#### Access to the AICSImage Object and Metadata\n\n![napari viewer with console open showing `viewer.layers[0].metadata`](https://raw.githubusercontent.com/AllenCellModeling/napari-aicsimageio/main/images/console-access.png)\n\nYou can access the `AICSImage` object used to load the image pixel data and\nimage metadata using the built-in napari console:\n\n```python\nimg = viewer.layers[0].metadata[\"aicsimage\"]\nimg.dims.order # TCZYX\nimg.channel_names # [\"Bright\", \"Struct\", \"Nuc\", \"Memb\"]\nimg.get_image_dask_data(\"ZYX\") # dask.array.Array\n```\n\nThe napari layer metadata dictionary also stores a shorthand\nfor the raw image metadata:\n\n```python\nviewer.layers[0].metadata[\"raw_image_metadata\"]\n```\n\nThe metadata is returned in whichever format is used by the underlying\nfile format reader, i.e. for CZI the raw metadata is returned as\nan `xml.etree.ElementTree.Element`, for OME-TIFF the raw metadata is returned\nas an `OME` object from `ome-types`.\n\nLastly, if the underlying file format reader has an OME metadata conversion function,\nyou may additionally see a key in the napari layer metadata dictionary\ncalled `\"ome_types\"`. For example, because the AICSImageIO\n`CZIReader` and `BioformatsReader` both support converting raw image metadata\nto OME metadata, you will see an `\"ome_types\"` key that stores the metadata transformed\ninto the OME metadata model.\n\n```python\nviewer.layers[0].metadata[\"ome_types\"] # OME object from ome-types\n```\n\n#### Mosaic Reading\n\nWhen reading CZI or LIF images, if the image is a mosaic tiled image, `napari-aicsimageio`\nwill return the reconstructed image:\n\n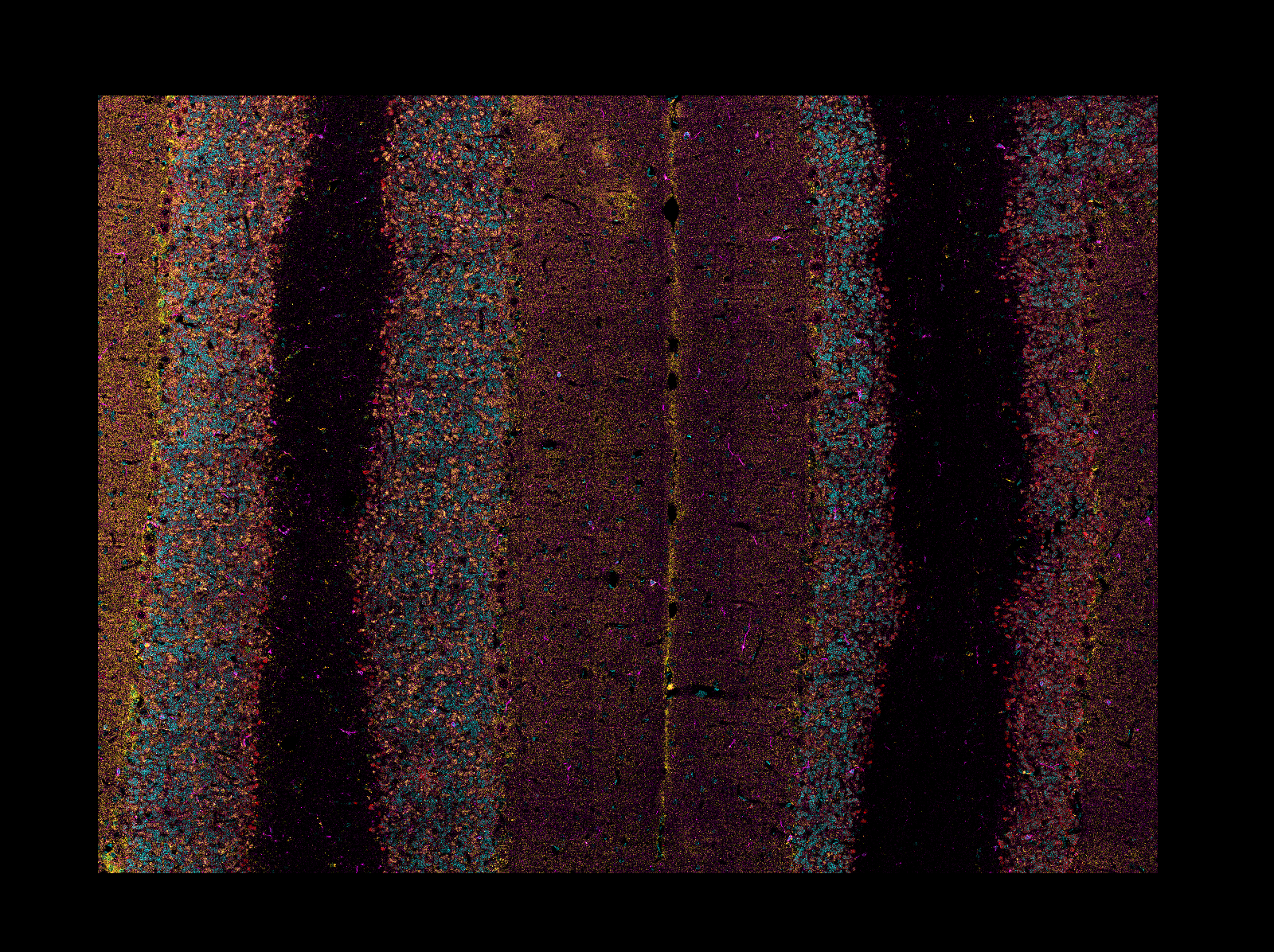\n\n## Development\n\nSee [CONTRIBUTING.md](CONTRIBUTING.md) for information related to developing the code.\n\nFor additional file format support, contributed directly to\n[AICSImageIO](https://github.com/AllenCellModeling/aicsimageio).\nNew file format support will become directly available in this\nplugin on new `aicsimageio` releases.\n\n## Citation\n\nIf you find `aicsimageio` _(or `napari-aicsimageio`)_ useful, please cite as:\n\n> AICSImageIO Contributors (2021). AICSImageIO: Image Reading, Metadata Conversion, and Image Writing for Microscopy Images in Pure Python [Computer software]. GitHub. https://github.com/AllenCellModeling/aicsimageio\n\n_Free software: GPLv3_\n",
"description_content_type": "text/markdown",
"keywords": "aicsimageio,CZI,image reading,imageio,LIF,metadata,napari,ND2,TIFF",
"home_page": null,
"download_url": null,
"author": null,
"author_email": "Eva Maxfield Brown <[email protected]>, Talley Lambert <[email protected]>",
"maintainer": null,
"maintainer_email": null,
"license": "GPLv3",
"classifier": [
"Development Status :: 5 - Production/Stable",
"Intended Audience :: Science/Research",
"Framework :: napari",
"License :: OSI Approved :: GNU General Public License v3 (GPLv3)",
"Operating System :: OS Independent",
"Programming Language :: Python :: 3",
"Programming Language :: Python :: 3.8",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
"Topic :: Scientific/Engineering :: Bio-Informatics",
"Topic :: Scientific/Engineering :: Image Processing",
"Topic :: Scientific/Engineering :: Information Analysis",
"Topic :: Scientific/Engineering :: Visualization",
"Topic :: Scientific/Engineering"
],
"requires_dist": [
"aicsimageio[all] (>=4.6.3)",
"fsspec[http] (>=2022.7.1)",
"napari (>=0.4.11)",
"psutil (>=5.7.0)",
"aicspylibczi (>=3.0.5)",
"bioformats-jar",
"readlif (>=0.6.4)",
"black (>=19.10b0) ; extra == 'dev'",
"coverage (>=5.1) ; extra == 'dev'",
"docutils (<0.16,>=0.10) ; extra == 'dev'",
"flake8-debugger (>=3.2.1) ; extra == 'dev'",
"flake8-pyprojecttoml ; extra == 'dev'",
"flake8 (>=3.8.3) ; extra == 'dev'",
"ipython (>=7.15.0) ; extra == 'dev'",
"isort (>=5.7.0) ; extra == 'dev'",
"mypy (>=0.800) ; extra == 'dev'",
"pytest-runner (>=5.2) ; extra == 'dev'",
"twine (>=3.1.1) ; extra == 'dev'",
"wheel (>=0.34.2) ; extra == 'dev'",
"PyQt5 ; extra == 'test'",
"pytest (>=5.4.3) ; extra == 'test'",
"pytest-qt (~=4.0) ; extra == 'test'",
"pytest-cov (>=2.9.0) ; extra == 'test'",
"pytest-raises (>=0.11) ; extra == 'test'",
"pytest-xvfb (~=2.0) ; extra == 'test'",
"quilt3 (~=3.4.0) ; extra == 'test'"
],
"requires_python": ">=3.8",
"requires_external": null,
"project_url": [
"Homepage, https://github.com/AllenCellModeling/napari-aicsimageio",
"Bug Tracker, https://github.com/AllenCellModeling/napari-aicsimageio/issues",
"Documentation, https://github.com/AllenCellModeling/napari-aicsimageio#README.md",
"User Support, https://github.com/AllenCellModeling/napari-aicsimageio/issues"
],
"provides_extra": [
"dev",
"test"
],
"provides_dist": null,
"obsoletes_dist": null
},
"npe1_shim": false
}