diff --git a/README.md b/README.md
index 36885689..62e3d326 100644
--- a/README.md
+++ b/README.md
@@ -11,12 +11,30 @@ The following methods are being developed:
- Image representation learning
- Self-supervised learning of the cell state and organelle phenotypes
-VisCy is currently considered alpha software and is under active development.
-Frequent breaking changes are expected.
+
+ Note:
+ VisCy is currently considered alpha software and is under active development. Frequent breaking changes are expected.
+
## Virtual staining
-
+### Pipeline
A full illustration of the virtual staining pipeline can be found [here](docs/virtual_staining.md).
+
+### Library of virtual staining (VS) models
+The robust virtual staining models (i.e *VSCyto2D*, *VSCyto3D*, *VSNeuromast*), and fine-tuned models can be found [here](https://github.com/mehta-lab/VisCy/wiki/Library-of-virtual-staining-(VS)-Models)
+
+### Demos
+#### Image-to-Image translation using VisCy
+- [Guide for Virtual Staining Models](https://github.com/mehta-lab/VisCy/wiki/virtual-staining-instructions):
+Instructions for how to train and run inference on ViSCy's virtual staining models (*VSCyto3D*, *VSCyto2D* and *VSNeuromast*)
+
+- [Image translation Exercise](./dlmbl_exercise/solution.py):
+Example showing how to use VisCy to train, predict and evaluate the VSCyto2D model. This notebook was developed for the [DL@MBL2024](https://github.com/dlmbl/DL-MBL-2024) course.
+
+- [Virtual staining exercise](./img2img_translation/solution.py): exploring the label-free to fluorescence virtual staining and florescence to label-free image translation task using VisCy UneXt2.
+More usage examples and demos can be found [here](https://github.com/mehta-lab/VisCy/blob/b7af9687c6409c738731ea47f66b74db2434443c/examples/virtual_staining/README.md)
+
+### Gallery
Below are some examples of virtually stained images (click to play videos).
See the full gallery [here](https://github.com/mehta-lab/VisCy/wiki/Gallery).
@@ -100,19 +118,14 @@ publisher = {eLife Sciences Publications, Ltd},
viscy --help
```
+## Contributing
For development installation, see [the contributing guide](CONTRIBUTING.md).
+## Additional Notes
The pipeline is built using the [PyTorch Lightning](https://www.pytorchlightning.ai/index.html) framework.
The [iohub](https://github.com/czbiohub-sf/iohub) library is used
for reading and writing data in [OME-Zarr](https://www.nature.com/articles/s41592-021-01326-w) format.
The full functionality is tested on Linux `x86_64` with NVIDIA Ampere GPUs (CUDA 12.4).
Some features (e.g. mixed precision and distributed training) may not be available with other setups,
-see [PyTorch documentation](https://pytorch.org) for details.
-
-### Demos
-Check out our demos for:
-- [Virtual staining](https://github.com/mehta-lab/VisCy/tree/main/examples/demos) - training, inference and evaluation
-
-### Library of virtual staining (VS) models
-The robust virtual staining models (i.e *VSCyto2D*, *VSCyto3D*, *VSNeuromast*), and fine-tuned models can be found [here](https://github.com/mehta-lab/VisCy/wiki/Library-of-virtual-staining-(VS)-Models)
\ No newline at end of file
+see [PyTorch documentation](https://pytorch.org) for details.
\ No newline at end of file
diff --git a/examples/demos/README.md b/examples/demos/README.md
deleted file mode 100644
index 3735caea..00000000
--- a/examples/demos/README.md
+++ /dev/null
@@ -1,29 +0,0 @@
-# VisCy usage examples
-
-Examples scripts showcasing the usage of VisCy.
-
-## Virtual staining
-
-### Training
-
-- WIP: DL@MBL notebooks
-
-### Inference
-
-- [Inference with VSCyto2D](./demo_vscyto2d.py):
-2D inference example on 20x A549 cell data. (Phase to nuclei and plasma membrane).
-- [Inference with VSCyto3D](./demos/demo_vscyto3d.py):
-3D inference example on 63x HEK293T cell data. (Phase to nuclei and plasma membrane).
-- [Inference VSNeuromast](./demo_vsneuromast.py):
-3D inference example of 63x zebrafish neuromast data (Phase to nuclei and plasma membrane)
-
-## Notes
-
-To run the examples, execute each individual script, for example:
-
-```sh
-python demo_vscyto2d.py
-```
-
-These scripts can also be ran interactively in many IDEs as notebooks,
-for example in VS Code, PyCharm, and Spyder.
diff --git a/examples/virtual_staining/README.md b/examples/virtual_staining/README.md
new file mode 100644
index 00000000..fec3c120
--- /dev/null
+++ b/examples/virtual_staining/README.md
@@ -0,0 +1,18 @@
+# VisCy usage examples
+
+Examples scripts showcasing the usage of VisCy for different computer vision tasks.
+
+## Virtual staining
+### Image-to-Image translation using VisCy
+- [Guide for Virtual Staining Models](https://github.com/mehta-lab/VisCy/wiki/virtual-staining-instructions):
+Instructions for how to train and run inference on ViSCy's virtual staining models (*VSCyto3D*, *VSCyto2D* and *VSNeuromast*)
+
+- [Image translation Exercise](./dlmbl_exercise/solution.py):
+Example showing how to use VisCy to train, predict and evaluate the VSCyto2D model. This notebook was developed for the [DL@MBL2024](https://github.com/dlmbl/DL-MBL-2024) course.
+
+- [Virtual staining exercise](./img2img_translation/solution.py): exploring the label-free to fluorescence virtual staining and florescence to label-free image translation task using VisCy UneXt2.
+
+## Notes
+To run the examples, make sure to activate the `viscy` environment. Follow the instructions for each demo.
+
+These scripts can also be ran interactively in many IDEs as notebooks,for example in VS Code, PyCharm, and Spyder.
diff --git a/examples/demos/demo_vscyto2d.py b/examples/virtual_staining/VS_model_inference/demo_vscyto2d.py
similarity index 95%
rename from examples/demos/demo_vscyto2d.py
rename to examples/virtual_staining/VS_model_inference/demo_vscyto2d.py
index c4430419..1c920a28 100644
--- a/examples/demos/demo_vscyto2d.py
+++ b/examples/virtual_staining/VS_model_inference/demo_vscyto2d.py
@@ -11,7 +11,6 @@
from iohub import open_ome_zarr
from plot import plot_vs_n_fluor
-
# Viscy classes for the trainer and model
from viscy.data.hcs import HCSDataModule
from viscy.light.engine import FcmaeUNet
@@ -31,13 +30,9 @@
root_dir = Path("")
# Download from
# https://public.czbiohub.org/comp.micro/viscy/VSCyto2D/test/a549_hoechst_cellmask_test.zarr/
-input_data_path = (
- root_dir / "VSCyto2D/test/a549_hoechst_cellmask_test.zarr"
-)
+input_data_path = root_dir / "VSCyto2D/test/a549_hoechst_cellmask_test.zarr"
# Download from GitHub release page of v0.1.0
-model_ckpt_path = (
- root_dir / "VisCy-0.1.0-VS-models/VSCyto2D/epoch=399-step=23200.ckpt"
-)
+model_ckpt_path = root_dir / "VisCy-0.1.0-VS-models/VSCyto2D/epoch=399-step=23200.ckpt"
# Zarr store to save the predictions
output_path = root_dir / "./a549_prediction.zarr"
# FOV of interest
diff --git a/examples/demos/demo_vscyto3d.py b/examples/virtual_staining/VS_model_inference/demo_vscyto3d.py
similarity index 97%
rename from examples/demos/demo_vscyto3d.py
rename to examples/virtual_staining/VS_model_inference/demo_vscyto3d.py
index eb4aff2d..928de1b7 100644
--- a/examples/demos/demo_vscyto3d.py
+++ b/examples/virtual_staining/VS_model_inference/demo_vscyto3d.py
@@ -11,9 +11,7 @@
from iohub import open_ome_zarr
from plot import plot_vs_n_fluor
-
from viscy.data.hcs import HCSDataModule
-
# Viscy classes for the trainer and model
from viscy.light.engine import VSUNet
from viscy.light.predict_writer import HCSPredictionWriter
@@ -30,7 +28,9 @@
# %%
# Download from
# https://public.czbiohub.org/comp.micro/viscy/VSCyto3D/test/no_pertubation_Phase1e-3_Denconv_Nuc8e-4_Mem8e-4_pad15_bg50.zarr/
-input_data_path = "VSCyto3D/test/no_pertubation_Phase1e-3_Denconv_Nuc8e-4_Mem8e-4_pad15_bg50.zarr"
+input_data_path = (
+ "VSCyto3D/test/no_pertubation_Phase1e-3_Denconv_Nuc8e-4_Mem8e-4_pad15_bg50.zarr"
+)
# Download from GitHub release page of v0.1.0
model_ckpt_path = "VisCy-0.1.0-VS-models/VSCyto3D/epoch=48-step=18130.ckpt"
# Zarr store to save the predictions
diff --git a/examples/demos/demo_vsneuromast.py b/examples/virtual_staining/VS_model_inference/demo_vsneuromast.py
similarity index 97%
rename from examples/demos/demo_vsneuromast.py
rename to examples/virtual_staining/VS_model_inference/demo_vsneuromast.py
index a24cb309..017ad4ef 100644
--- a/examples/demos/demo_vsneuromast.py
+++ b/examples/virtual_staining/VS_model_inference/demo_vsneuromast.py
@@ -11,9 +11,7 @@
from iohub import open_ome_zarr
from plot import plot_vs_n_fluor
-
from viscy.data.hcs import HCSDataModule
-
# Viscy classes for the trainer and model
from viscy.light.engine import VSUNet
from viscy.light.predict_writer import HCSPredictionWriter
@@ -30,7 +28,9 @@
# %%
# Download from
# https://public.czbiohub.org/comp.micro/viscy/VSNeuromast/test/20230803_fish2_60x_1_cropped_zyx_resampled_clipped_2.zarr/
-input_data_path = "VSNeuromast/test/20230803_fish2_60x_1_cropped_zyx_resampled_clipped_2.zarr"
+input_data_path = (
+ "VSNeuromast/test/20230803_fish2_60x_1_cropped_zyx_resampled_clipped_2.zarr"
+)
# Download from GitHub release page of v0.1.0
model_ckpt_path = "VisCy-0.1.0-VS-models/VSNeuromast/timelapse_finetine_1hr_dT_downsample_lr1e-4_45epoch_clahe_v5/epoch=44-step=1215.ckpt"
# Zarr store to save the predictions
diff --git a/examples/demos/plot.py b/examples/virtual_staining/VS_model_inference/plot.py
similarity index 100%
rename from examples/demos/plot.py
rename to examples/virtual_staining/VS_model_inference/plot.py
diff --git a/examples/virtual_staining/dlmbl_exercise/README.md b/examples/virtual_staining/dlmbl_exercise/README.md
new file mode 100644
index 00000000..eeababb2
--- /dev/null
+++ b/examples/virtual_staining/dlmbl_exercise/README.md
@@ -0,0 +1,88 @@
+# Exercise 6: Image translation - Part 1
+
+This demo script was developed for the DL@MBL 2024 course by Eduardo Hirata-Miyasaki, Ziwen Liu and Shalin Mehta, with many inputs and bugfixes by [Morgan Schwartz](https://github.com/msschwartz21), [Caroline Malin-Mayor](https://github.com/cmalinmayor), and [Peter Park](https://github.com/peterhpark).
+
+
+# Image translation (Virtual Staining)
+
+Written by Eduardo Hirata-Miyasaki, Ziwen Liu, and Shalin Mehta, CZ Biohub San Francisco.
+
+## Overview
+
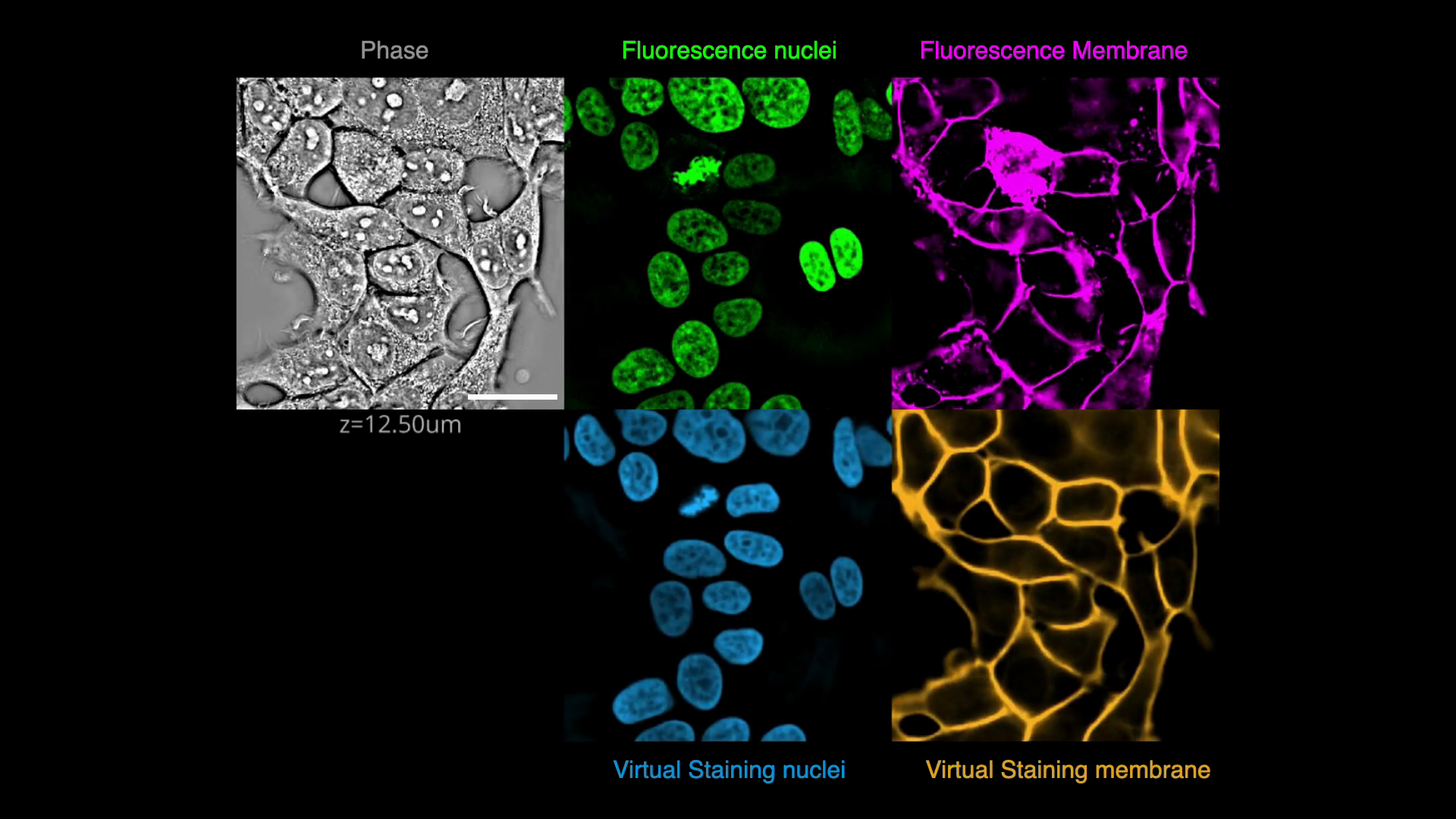
+In this exercise, we will predict fluorescence images of nuclei and plasma membrane markers from quantitative phase images of cells, i.e., we will _virtually stain_ the nuclei and plasma membrane visible in the phase image.
+This is an example of an image translation task. We will apply spatial and intensity augmentations to train robust models and evaluate their performance. Finally, we will explore the opposite process of predicting a phase image from a fluorescence membrane label.
+
+[](https://github.com/mehta-lab/VisCy/assets/67518483/d53a81eb-eb37-44f3-b522-8bd7bddc7755)
+(Click on image to play video)
+
+## Goals
+
+### Part 1: Learn to use iohub (I/O library), VisCy dataloaders, and TensorBoard.
+
+ - Use a OME-Zarr dataset of 34 FOVs of adenocarcinomic human alveolar basal epithelial cells (A549),
+ each FOV has 3 channels (phase, nuclei, and cell membrane).
+ The nuclei were stained with DAPI and the cell membrane with Cellmask.
+ - Explore OME-Zarr using [iohub](https://czbiohub-sf.github.io/iohub/main/index.html)
+ and the high-content-screen (HCS) format.
+ - Use [MONAI](https://monai.io/) to implement data augmentations.
+
+### Part 2: Train and evaluate the model to translate phase into fluorescence, and vice versa.
+ - Train a 2D UNeXt2 model to predict nuclei and membrane from phase images.
+ - Compare the performance of the trained model and a pre-trained model.
+ - Evaluate the model using pixel-level and instance-level metrics.
+
+
+Checkout [VisCy](https://github.com/mehta-lab/VisCy/tree/main/examples/demos),
+our deep learning pipeline for training and deploying computer vision models
+for image-based phenotyping including the robust virtual staining of landmark organelles.
+VisCy exploits recent advances in data and metadata formats
+([OME-zarr](https://www.nature.com/articles/s41592-021-01326-w)) and DL frameworks,
+[PyTorch Lightning](https://lightning.ai/) and [MONAI](https://monai.io/).
+
+## Setup
+
+Make sure that you are inside of the `image_translation` folder by using the `cd` command to change directories if needed.
+
+Make sure that you can use conda to switch environments.
+
+```bash
+conda init
+```
+
+**Close your shell, and login again.**
+
+Run the setup script to create the environment for this exercise and download the dataset.
+```bash
+sh setup.sh
+```
+Activate your environment
+```bash
+conda activate 06_image_translation
+```
+
+## Use vscode
+
+Install vscode, install jupyter extension inside vscode, and setup [cell mode](https://code.visualstudio.com/docs/python/jupyter-support-py). Open [solution.py](solution.py) and run the script interactively.
+
+## Use Jupyter Notebook
+
+The matching exercise and solution notebooks can be found [here](https://github.com/dlmbl/image_translation/tree/28e0e515b4a8ad3f392a69c8341e105f730d204f) on the course repository.
+
+Launch a jupyter environment
+
+```
+jupyter notebook
+```
+
+...and continue with the instructions in the notebook.
+
+If `06_image_translation` is not available as a kernel in jupyter, run:
+
+```
+python -m ipykernel install --user --name=06_image_translation
+```
+
+### References
+
+- [Liu, Z. and Hirata-Miyasaki, E. et al. (2024) Robust Virtual Staining of Cellular Landmarks](https://www.biorxiv.org/content/10.1101/2024.05.31.596901v2.full.pdf)
+- [Guo et al. (2020) Revealing architectural order with quantitative label-free imaging and deep learning. eLife](https://elifesciences.org/articles/55502)
diff --git a/examples/demo_dlmbl/convert-solution.py b/examples/virtual_staining/dlmbl_exercise/convert-solution.py
similarity index 90%
rename from examples/demo_dlmbl/convert-solution.py
rename to examples/virtual_staining/dlmbl_exercise/convert-solution.py
index 91d7e46c..206ea3d0 100644
--- a/examples/demo_dlmbl/convert-solution.py
+++ b/examples/virtual_staining/dlmbl_exercise/convert-solution.py
@@ -1,7 +1,8 @@
import argparse
from nbconvert.exporters import NotebookExporter
-from nbconvert.preprocessors import ClearOutputPreprocessor, TagRemovePreprocessor
+from nbconvert.preprocessors import (ClearOutputPreprocessor,
+ TagRemovePreprocessor)
from traitlets.config import Config
diff --git a/examples/demo_dlmbl/prepare-exercise.sh b/examples/virtual_staining/dlmbl_exercise/prepare-exercise.sh
similarity index 100%
rename from examples/demo_dlmbl/prepare-exercise.sh
rename to examples/virtual_staining/dlmbl_exercise/prepare-exercise.sh
diff --git a/examples/virtual_staining/dlmbl_exercise/setup.sh b/examples/virtual_staining/dlmbl_exercise/setup.sh
new file mode 100644
index 00000000..e185c4d2
--- /dev/null
+++ b/examples/virtual_staining/dlmbl_exercise/setup.sh
@@ -0,0 +1,32 @@
+#!/usr/bin/env -S bash -i
+
+START_DIR=$(pwd)
+
+# Create conda environment
+conda create -y --name 06_image_translation python=3.10
+
+# Install ipykernel in the environment.
+conda install -y ipykernel nbformat nbconvert black jupytext ipywidgets --name 06_image_translation
+# Specifying the environment explicitly.
+# conda activate sometimes doesn't work from within shell scripts.
+
+# install viscy and its dependencies`s in the environment using pip.
+# Find path to the environment - conda activate doesn't work from within shell scripts.
+ENV_PATH=$(conda info --envs | grep 06_image_translation | awk '{print $NF}')
+$ENV_PATH/bin/pip install "viscy[metrics,visual]==0.2.0"
+
+# Create the directory structure
+mkdir -p ~/data/06_image_translation/training
+mkdir -p ~/data/06_image_translation/test
+mkdir -p ~/data/06_image_translation/pretrained_models
+# Change to the target directory
+cd ~/data/06_image_translation/training
+# Download the OME-Zarr dataset recursively
+wget -m -np -nH --cut-dirs=5 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VS_datasets/VSCyto2D/training/a549_hoechst_cellmask_train_val.zarr/"
+cd ~/data/06_image_translation/test
+wget -m -np -nH --cut-dirs=5 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VS_datasets/VSCyto2D/test/a549_hoechst_cellmask_test.zarr/"
+cd ~/data/06_image_translation/pretrained_models
+wget -m -np -nH --cut-dirs=5 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VS_models/VSCyto2D/VSCyto2D/epoch=399-step=23200.ckpt"
+
+# Change back to the starting directory
+cd $START_DIR
diff --git a/examples/virtual_staining/dlmbl_exercise/solution.py b/examples/virtual_staining/dlmbl_exercise/solution.py
new file mode 100644
index 00000000..93fc4917
--- /dev/null
+++ b/examples/virtual_staining/dlmbl_exercise/solution.py
@@ -0,0 +1,1326 @@
+# %% [markdown]
+"""
+# Image translation (Virtual Staining - Part 1
+
+Written by Eduardo Hirata-Miyasaki, Ziwen Liu, and Shalin Mehta, CZ Biohub San Francisco.
+
+## Overview
+
+In this exercise, we will predict fluorescence images of
+nuclei and plasma membrane markers from quantitative phase images of cells,
+i.e., we will _virtually stain_ the nuclei and plasma membrane
+visible in the phase image.
+This is an example of an image translation task.
+We will apply spatial and intensity augmentations to train robust models
+and evaluate their performance using a regression approach.
+
+[](https://github.com/mehta-lab/VisCy/assets/67518483/d53a81eb-eb37-44f3-b522-8bd7bddc7755)
+(Click on image to play video)
+"""
+
+# %% [markdown]
+"""
+### Goals
+
+#### Part 1: Learn to use iohub (I/O library), VisCy dataloaders, and TensorBoard.
+
+ - Use a OME-Zarr dataset of 34 FOVs of adenocarcinomic human alveolar basal epithelial cells (A549),
+ each FOV has 3 channels (phase, nuclei, and cell membrane).
+ The nuclei were stained with DAPI and the cell membrane with Cellmask.
+ - Explore OME-Zarr using [iohub](https://czbiohub-sf.github.io/iohub/main/index.html)
+ and the high-content-screen (HCS) format.
+ - Use [MONAI](https://monai.io/) to implement data augmentations.
+
+#### Part 2: Train and evaluate the model to translate phase into fluorescence.
+ - Train a 2D UNeXt2 model to predict nuclei and membrane from phase images.
+ - Compare the performance of the trained model and a pre-trained model.
+ - Evaluate the model using pixel-level and instance-level metrics.
+
+
+Checkout [VisCy](https://github.com/mehta-lab/VisCy/tree/main/examples/demos),
+our deep learning pipeline for training and deploying computer vision models
+for image-based phenotyping including the robust virtual staining of landmark organelles.
+VisCy exploits recent advances in data and metadata formats
+([OME-zarr](https://www.nature.com/articles/s41592-021-01326-w)) and DL frameworks,
+[PyTorch Lightning](https://lightning.ai/) and [MONAI](https://monai.io/).
+
+### References
+
+- [Liu, Z. and Hirata-Miyasaki, E. et al. (2024) Robust Virtual Staining of Cellular Landmarks](https://www.biorxiv.org/content/10.1101/2024.05.31.596901v2.full.pdf)
+- [Guo et al. (2020) Revealing architectural order with quantitative label-free imaging and deep learning. eLife](https://elifesciences.org/articles/55502)
+"""
+
+
+# %% [markdown]
+"""
+📖 As you work through parts 2, please share the layouts of your models (output of torchview)
+and their performance with everyone via
+[this Google Doc](https://docs.google.com/document/d/1Mq-yV8FTG02xE46Mii2vzPJVYSRNdeOXkeU-EKu-irE/edit?usp=sharing). 📖
+"""
+# %% [markdown]
+"""
+
+The exercise is organized in 2 parts
+
+
+- Part 1 - Learn to use iohub (I/O library), VisCy dataloaders, and tensorboard.
+- Part 2 - Train and evaluate the model to translate phase into fluorescence.
+
+
+
+Set your python kernel to 06_image_translation
+
+"""
+# %% [markdown]
+"""
+## Part 1: Log training data to tensorboard, start training a model.
+---------
+Learning goals:
+
+- Load the OME-zarr dataset and examine the channels (A549).
+- Configure and understand the data loader.
+- Log some patches to tensorboard.
+- Initialize a 2D UNeXt2 model for virtual staining of nuclei and membrane from phase.
+- Start training the model to predict nuclei and membrane from phase.
+"""
+
+# %% Imports
+import os
+from glob import glob
+from pathlib import Path
+from typing import Tuple
+
+import matplotlib.pyplot as plt
+import numpy as np
+import pandas as pd
+import torch
+import torchview
+import torchvision
+from cellpose import models
+from iohub import open_ome_zarr
+from iohub.reader import print_info
+from lightning.pytorch import seed_everything
+from lightning.pytorch.loggers import TensorBoardLogger
+from natsort import natsorted
+from numpy.typing import ArrayLike
+from skimage import metrics # for metrics.
+# pytorch lightning wrapper for Tensorboard.
+from skimage.color import label2rgb
+from torch.utils.tensorboard import SummaryWriter # for logging to tensorboard
+from torchmetrics.functional import accuracy, dice, jaccard_index
+from tqdm import tqdm
+# HCSDataModule makes it easy to load data during training.
+from viscy.data.hcs import HCSDataModule
+from viscy.evaluation.evaluation_metrics import mean_average_precision
+# Trainer class and UNet.
+from viscy.light.engine import MixedLoss, VSUNet
+from viscy.light.trainer import VSTrainer
+# training augmentations
+from viscy.transforms import (NormalizeSampled, RandAdjustContrastd,
+ RandAffined, RandGaussianNoised,
+ RandGaussianSmoothd, RandScaleIntensityd,
+ RandWeightedCropd)
+
+# %%
+# seed random number generators for reproducibility.
+seed_everything(42, workers=True)
+
+# Paths to data and log directory
+top_dir = Path(
+ f"/hpc/mydata/{os.environ['USER']}/data/"
+) # TODO: Change this to point to your data directory.
+
+data_path = (
+ top_dir / "06_image_translation/training/a549_hoechst_cellmask_train_val.zarr"
+)
+log_dir = top_dir / "06_image_translation/logs/"
+
+if not data_path.exists():
+ raise FileNotFoundError(
+ f"Data not found at {data_path}. Please check the top_dir and data_path variables."
+ )
+
+# %%
+# Create log directory if needed, and launch tensorboard
+log_dir.mkdir(parents=True, exist_ok=True)
+
+# %% [markdown] tags=[]
+"""
+The next cell starts tensorboard.
+
+
+If you launched jupyter lab from ssh terminal, add --host <your-server-name> to the tensorboard command below. <your-server-name> is the address of your compute node that ends in amazonaws.com.
+
+
+
+
+If you are using VSCode and a remote server, you will need to forward the port to view the tensorboard.
+Take note of the port number was assigned in the previous cell.(i.e
http://localhost:{port_number_assigned})
+
+Locate the your VSCode terminal and select the
Ports tab
+
+- Add a new port with the
port_number_assigned
+
+Click on the link to view the tensorboard and it should open in your browser.
+
+You can inspect the tree structure by using your terminal:
+ iohub info -v "path-to-ome-zarr"
+
+
+More info on the CLI:
+iohub info --help to see the help menu.
+
+"""
+# %%
+# This is the python function called by `iohub info` CLI command
+print_info(data_path, verbose=True)
+
+# Open and inspect the dataset.
+dataset = open_ome_zarr(data_path)
+
+# %%
+# Use the field and pyramid_level below to visualize data.
+row = 0
+col = 0
+field = 9 # TODO: Change this to explore data.
+
+
+# NOTE: this dataset only has one level
+pyaramid_level = 0
+
+# `channel_names` is the metadata that is stored with data according to the OME-NGFF spec.
+n_channels = len(dataset.channel_names)
+
+image = dataset[f"{row}/{col}/{field}/{pyaramid_level}"].numpy()
+print(f"data shape: {image.shape}, FOV: {field}, pyramid level: {pyaramid_level}")
+
+figure, axes = plt.subplots(1, n_channels, figsize=(9, 3))
+
+for i in range(n_channels):
+ for i in range(n_channels):
+ channel_image = image[0, i, 0]
+ # Adjust contrast to 0.5th and 99.5th percentile of pixel values.
+ p_low, p_high = np.percentile(channel_image, (0.5, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[i].imshow(channel_image, cmap="gray")
+ axes[i].axis("off")
+ axes[i].set_title(dataset.channel_names[i])
+plt.tight_layout()
+
+# %% [markdown]
+#
+#
+# ### Task 1.1
+#
+# Look at a couple different fields of view by changing the value in the cell above.
+# Check the cell density, the cell morphologies, and fluorescence signal.
+#
+
+# %% [markdown]
+"""
+## Explore the effects of augmentation on batch.
+
+VisCy builds on top of PyTorch Lightning. PyTorch Lightning is a thin wrapper around PyTorch that allows rapid experimentation. It provides a [DataModule](https://lightning.ai/docs/pytorch/stable/data/datamodule.html) to handle loading and processing of data during training. VisCy provides a child class, `HCSDataModule` to make it intuitve to access data stored in the HCS layout.
+
+The dataloader in `HCSDataModule` returns a batch of samples. A `batch` is a list of dictionaries. The length of the list is equal to the batch size. Each dictionary consists of following key-value pairs.
+- `source`: the input image, a tensor of size 1*1*Y*X
+- `target`: the target image, a tensor of size 2*1*Y*X
+- `index` : the tuple of (location of field in HCS layout, time, and z-slice) of the sample.
+"""
+
+# %% [markdown]
+#
+#
+# ### Task 1.2
+#
+# Setup the data loader and log several batches to tensorboard.
+#
+# Based on the tensorboard images, what are the two channels in the target image?
+#
+# Note: If tensorboard is not showing images, try refreshing and using the "Images" tab.
+#
+
+# %%
+# Define a function to write a batch to tensorboard log.
+
+
+def log_batch_tensorboard(batch, batchno, writer, card_name):
+ """
+ Logs a batch of images to TensorBoard.
+
+ Args:
+ batch (dict): A dictionary containing the batch of images to be logged.
+ writer (SummaryWriter): A TensorBoard SummaryWriter object.
+ card_name (str): The name of the card to be displayed in TensorBoard.
+
+ Returns:
+ None
+ """
+ batch_phase = batch["source"][:, :, 0, :, :] # batch_size x z_size x Y x X tensor.
+ batch_membrane = batch["target"][:, 1, 0, :, :].unsqueeze(
+ 1
+ ) # batch_size x 1 x Y x X tensor.
+ batch_nuclei = batch["target"][:, 0, 0, :, :].unsqueeze(
+ 1
+ ) # batch_size x 1 x Y x X tensor.
+
+ p1, p99 = np.percentile(batch_membrane, (0.1, 99.9))
+ batch_membrane = np.clip((batch_membrane - p1) / (p99 - p1), 0, 1)
+
+ p1, p99 = np.percentile(batch_nuclei, (0.1, 99.9))
+ batch_nuclei = np.clip((batch_nuclei - p1) / (p99 - p1), 0, 1)
+
+ p1, p99 = np.percentile(batch_phase, (0.1, 99.9))
+ batch_phase = np.clip((batch_phase - p1) / (p99 - p1), 0, 1)
+
+ [N, C, H, W] = batch_phase.shape
+ interleaved_images = torch.zeros((3 * N, C, H, W), dtype=batch_phase.dtype)
+ interleaved_images[0::3, :] = batch_phase
+ interleaved_images[1::3, :] = batch_nuclei
+ interleaved_images[2::3, :] = batch_membrane
+
+ grid = torchvision.utils.make_grid(interleaved_images, nrow=3)
+
+ # add the grid to tensorboard
+ writer.add_image(card_name, grid, batchno)
+
+
+# %%
+# Define a function to visualize a batch on jupyter, in case tensorboard is finicky
+def log_batch_jupyter(batch):
+ """
+ Logs a batch of images on jupyter using ipywidget.
+
+ Args:
+ batch (dict): A dictionary containing the batch of images to be logged.
+
+ Returns:
+ None
+ """
+ batch_phase = batch["source"][:, :, 0, :, :] # batch_size x z_size x Y x X tensor.
+ batch_size = batch_phase.shape[0]
+ batch_membrane = batch["target"][:, 1, 0, :, :].unsqueeze(
+ 1
+ ) # batch_size x 1 x Y x X tensor.
+ batch_nuclei = batch["target"][:, 0, 0, :, :].unsqueeze(
+ 1
+ ) # batch_size x 1 x Y x X tensor.
+
+ p1, p99 = np.percentile(batch_membrane, (0.1, 99.9))
+ batch_membrane = np.clip((batch_membrane - p1) / (p99 - p1), 0, 1)
+
+ p1, p99 = np.percentile(batch_nuclei, (0.1, 99.9))
+ batch_nuclei = np.clip((batch_nuclei - p1) / (p99 - p1), 0, 1)
+
+ p1, p99 = np.percentile(batch_phase, (0.1, 99.9))
+ batch_phase = np.clip((batch_phase - p1) / (p99 - p1), 0, 1)
+
+ plt.figure()
+ fig, axes = plt.subplots(
+ batch_size, n_channels, figsize=(n_channels * 2, batch_size * 2)
+ )
+ [N, C, H, W] = batch_phase.shape
+ for sample_id in range(batch_size):
+ axes[sample_id, 0].imshow(batch_phase[sample_id, 0])
+ axes[sample_id, 1].imshow(batch_nuclei[sample_id, 0])
+ axes[sample_id, 2].imshow(batch_membrane[sample_id, 0])
+
+ for i in range(n_channels):
+ axes[sample_id, i].axis("off")
+ axes[sample_id, i].set_title(dataset.channel_names[i])
+ plt.tight_layout()
+ plt.show()
+
+
+# %%
+# Initialize the data module.
+
+BATCH_SIZE = 4
+
+# 5 is a perfectly reasonable batch size
+# (batch size does not have to be a power of 2)
+# See: https://sebastianraschka.com/blog/2022/batch-size-2.html
+
+data_module = HCSDataModule(
+ data_path,
+ z_window_size=1,
+ architecture="UNeXt2_2D",
+ source_channel=["Phase3D"],
+ target_channel=["Nucl", "Mem"],
+ split_ratio=0.8,
+ batch_size=BATCH_SIZE,
+ num_workers=8,
+ yx_patch_size=(256, 256), # larger patch size makes it easy to see augmentations.
+ augmentations=[], # Turn off augmentation for now.
+ normalizations=[], # Turn off normalization for now.
+)
+data_module.setup("fit")
+
+print(
+ f"Samples in training set: {len(data_module.train_dataset)}, "
+ f"samples in validation set:{len(data_module.val_dataset)}"
+)
+train_dataloader = data_module.train_dataloader()
+
+# Instantiate the tensorboard SummaryWriter, logs the first batch and then iterates through all the batches and logs them to tensorboard.
+
+writer = SummaryWriter(log_dir=f"{log_dir}/view_batch")
+# Draw a batch and write to tensorboard.
+batch = next(iter(train_dataloader))
+log_batch_tensorboard(batch, 0, writer, "augmentation/none")
+writer.close()
+
+
+# %% [markdown]
+# If your tensorboard is causing issues, you can visualize directly on Jupyter /VSCode
+
+# %%
+log_batch_jupyter(batch)
+
+# %% [markdown] tags=[]
+#
+#
+# ### Task 1.3
+# Add augmentations to the datamodule and rerun the setup.
+#
+# What kind of augmentations do you think are important for this task?
+#
+# How do they make the model more robust?
+#
+# Add augmentations to rotate about $\pi$ around z-axis, 30% scale in y,x,
+# shearing of 10% and no padding with zeros with a probablity of 80%.
+#
+# Add a Gaussian noise with a mean of 0.0 and standard deviation of 0.3 with a probability of 50%.
+#
+# HINT: `RandAffined()` and `RandGaussianNoised()` are from
+# `viscy.transforms` [here](https://github.com/mehta-lab/VisCy/blob/main/viscy/transforms.py).
+# *Note these are MONAI transforms that have been redefined for VisCy.*
+# Can you tell what augmentation were applied from looking at the augmented images in Tensorboard?
+#
+# HINT:
+# [Compare your choice of augmentations by dowloading the pretrained models and config files](https://github.com/mehta-lab/VisCy/releases/download/v0.1.0/VisCy-0.1.0-VS-models.zip).
+#
+# %%
+# Here we turn on data augmentation and rerun setup
+source_channel = ["Phase3D"]
+target_channel = ["Nucl", "Mem"]
+
+augmentations = [
+ RandWeightedCropd(
+ keys=source_channel + target_channel,
+ spatial_size=(1, 256, 256),
+ num_samples=2,
+ w_key=target_channel[0],
+ ),
+ RandAdjustContrastd(keys=source_channel, prob=0.5, gamma=(0.8, 1.2)),
+ RandScaleIntensityd(keys=source_channel, factors=0.5, prob=0.5),
+ RandGaussianSmoothd(
+ keys=source_channel,
+ sigma_x=(0.25, 0.75),
+ sigma_y=(0.25, 0.75),
+ sigma_z=(0.0, 0.0),
+ prob=0.5,
+ ),
+ # #######################
+ # ##### TODO ########
+ # #######################
+ ##TODO: Add Random Affine Transorms
+ ## Write code below
+ ## TODO: Add Random Gaussian Noise
+ ## Write code below
+]
+
+normalizations = [
+ NormalizeSampled(
+ keys=source_channel + target_channel,
+ level="fov_statistics",
+ subtrahend="mean",
+ divisor="std",
+ )
+]
+
+data_module.augmentations = augmentations
+data_module.setup("fit")
+
+# get the new data loader with augmentation turned on
+augmented_train_dataloader = data_module.train_dataloader()
+
+# Draw batches and write to tensorboard
+writer = SummaryWriter(log_dir=f"{log_dir}/view_batch")
+augmented_batch = next(iter(augmented_train_dataloader))
+log_batch_tensorboard(augmented_batch, 0, writer, "augmentation/some")
+writer.close()
+
+# %% tags=["solution"]
+# #######################
+# ##### SOLUTION ########
+# #######################
+source_channel = ["Phase3D"]
+target_channel = ["Nucl", "Mem"]
+
+augmentations = [
+ RandWeightedCropd(
+ keys=source_channel + target_channel,
+ spatial_size=(1, 384, 384),
+ num_samples=2,
+ w_key=target_channel[0],
+ ),
+ RandAffined(
+ keys=source_channel + target_channel,
+ rotate_range=[3.14, 0.0, 0.0],
+ scale_range=[0.0, 0.3, 0.3],
+ prob=0.8,
+ padding_mode="zeros",
+ shear_range=[0.0, 0.01, 0.01],
+ ),
+ RandAdjustContrastd(keys=source_channel, prob=0.5, gamma=(0.8, 1.2)),
+ RandScaleIntensityd(keys=source_channel, factors=0.5, prob=0.5),
+ RandGaussianNoised(keys=source_channel, prob=0.5, mean=0.0, std=0.3),
+ RandGaussianSmoothd(
+ keys=source_channel,
+ sigma_x=(0.25, 0.75),
+ sigma_y=(0.25, 0.75),
+ sigma_z=(0.0, 0.0),
+ prob=0.5,
+ ),
+]
+
+normalizations = [
+ NormalizeSampled(
+ keys=source_channel + target_channel,
+ level="fov_statistics",
+ subtrahend="mean",
+ divisor="std",
+ )
+]
+
+data_module.augmentations = augmentations
+data_module.setup("fit")
+
+# get the new data loader with augmentation turned on
+augmented_train_dataloader = data_module.train_dataloader()
+
+# Draw batches and write to tensorboard
+writer = SummaryWriter(log_dir=f"{log_dir}/view_batch")
+augmented_batch = next(iter(augmented_train_dataloader))
+log_batch_tensorboard(augmented_batch, 0, writer, "augmentation/some")
+writer.close()
+
+
+# %% [markdown]
+# Visualize directly on Jupyter
+
+# %%
+log_batch_jupyter(augmented_batch)
+
+# %% [markdown]
+"""
+## Train a 2D U-Net model to predict nuclei and membrane from phase.
+
+### Construct a 2D UNeXt2 using VisCy
+See ``viscy.unet.networks.Unet2D.Unet2d`` ([source code](https://github.com/mehta-lab/VisCy/blob/7c5e4c1d68e70163cf514d22c475da8ea7dc3a88/viscy/unet/networks/Unet2D.py#L7)) for configuration details.
+"""
+# %%
+# Create a 2D UNet.
+GPU_ID = 0
+
+BATCH_SIZE = 12
+YX_PATCH_SIZE = (256, 256)
+
+# Dictionary that specifies key parameters of the model.
+
+phase2fluor_config = dict(
+ in_channels=1,
+ out_channels=2,
+ encoder_blocks=[3, 3, 9, 3],
+ dims=[96, 192, 384, 768],
+ decoder_conv_blocks=2,
+ stem_kernel_size=(1, 2, 2),
+ in_stack_depth=1,
+ pretraining=False,
+)
+
+phase2fluor_model = VSUNet(
+ architecture="UNeXt2_2D", # 2D UNeXt2 architecture
+ model_config=phase2fluor_config.copy(),
+ loss_function=MixedLoss(l1_alpha=0.5, l2_alpha=0.0, ms_dssim_alpha=0.5),
+ schedule="WarmupCosine",
+ lr=2e-4,
+ log_batches_per_epoch=5, # Number of samples from each batch to log to tensorboard.
+ freeze_encoder=False,
+)
+
+# %% [markdown]
+"""
+### Instantiate data module and trainer, test that we are setup to launch training.
+"""
+# %%
+source_channel = ["Phase3D"]
+target_channel = ["Nucl", "Mem"]
+# Setup the data module.
+phase2fluor_2D_data = HCSDataModule(
+ data_path,
+ architecture="UNeXt2_2D",
+ source_channel=source_channel,
+ target_channel=target_channel,
+ z_window_size=1,
+ split_ratio=0.8,
+ batch_size=BATCH_SIZE,
+ num_workers=8,
+ yx_patch_size=YX_PATCH_SIZE,
+ augmentations=augmentations,
+ normalizations=normalizations,
+)
+phase2fluor_2D_data.setup("fit")
+# fast_dev_run runs a single batch of data through the model to check for errors.
+trainer = VSTrainer(accelerator="gpu", devices=[GPU_ID], fast_dev_run=True)
+
+# trainer class takes the model and the data module as inputs.
+trainer.fit(phase2fluor_model, datamodule=phase2fluor_2D_data)
+
+
+# %% [markdown]
+# ## View model graph.
+#
+# PyTorch uses dynamic graphs under the hood.
+# The graphs are constructed on the fly.
+# This is in contrast to TensorFlow,
+# where the graph is constructed before the training loop and remains static.
+# In other words, the graph of the network can change with every forward pass.
+# Therefore, we need to supply an input tensor to construct the graph.
+# The input tensor can be a random tensor of the correct shape and type.
+# We can also supply a real image from the dataset.
+# The latter is more useful for debugging.
+
+# %% [markdown]
+#
+#
+# ### Task 1.4
+# Run the next cell to generate a graph representation of the model architecture.
+# Can you recognize the UNet structure and skip connections in this graph visualization?
+#
+
+# %%
+# visualize graph of phase2fluor model as image.
+model_graph_phase2fluor = torchview.draw_graph(
+ phase2fluor_model,
+ phase2fluor_2D_data.train_dataset[0]["source"][0].unsqueeze(dim=0),
+ roll=True,
+ depth=3, # adjust depth to zoom in.
+ device="cpu",
+ # expand_nested=True,
+)
+# Print the image of the model.
+model_graph_phase2fluor.visual_graph
+
+
+# %% [markdown]
+"""
+
+
+
Task 1.5
+Start training by running the following cell. Check the new logs on the tensorboard.
+
+"""
+
+# %%
+# Check if GPU is available
+# You can check by typing `nvidia-smi`
+GPU_ID = 0
+
+n_samples = len(phase2fluor_2D_data.train_dataset)
+steps_per_epoch = n_samples // BATCH_SIZE # steps per epoch.
+n_epochs = 50 # Set this to 50 or the number of epochs you want to train for.
+
+trainer = VSTrainer(
+ accelerator="gpu",
+ devices=[GPU_ID],
+ max_epochs=n_epochs,
+ log_every_n_steps=steps_per_epoch // 2,
+ # log losses and image samples 2 times per epoch.
+ logger=TensorBoardLogger(
+ save_dir=log_dir,
+ # lightning trainer transparently saves logs and model checkpoints in this directory.
+ name="phase2fluor",
+ log_graph=True,
+ ),
+)
+# Launch training and check that loss and images are being logged on tensorboard.
+trainer.fit(phase2fluor_model, datamodule=phase2fluor_2D_data)
+
+# %% [markdown]
+"""
+
+
+
Checkpoint 1
+
+While your model is training, let's think about the following questions:
+
+- What is the information content of each channel in the dataset?
+- How would you use image translation models?
+- What can you try to improve the performance of each model?
+
+
+Now the training has started,
+we can come back after a while and evaluate the performance!
+
+
+
+
Task 2.1 Define metrics
+
+For each of the above metrics, write a brief definition of what they are and what they mean
+for this image translation task. Use your favorite search engine and/or resources.
+
+
+"""
+
+# %% [markdown]
+# ```
+# #######################
+# ##### Todo ############
+# #######################
+#
+# ```
+#
+# - Pearson Correlation:
+#
+# - Structural similarity:
+
+# %% [markdown]
+"""
+Let's compute metrics directly and plot below.
+"""
+# %%
+# Setup the test data module.
+test_data_path = top_dir / "06_image_translation/test/a549_hoechst_cellmask_test.zarr"
+source_channel = ["Phase3D"]
+target_channel = ["Nucl", "Mem"]
+
+test_data = HCSDataModule(
+ test_data_path,
+ source_channel=source_channel,
+ target_channel=target_channel,
+ z_window_size=1,
+ batch_size=1,
+ num_workers=8,
+ architecture="UNeXt2",
+)
+test_data.setup("test")
+
+test_metrics = pd.DataFrame(
+ columns=["pearson_nuc", "SSIM_nuc", "pearson_mem", "SSIM_mem"]
+)
+
+# %% Compute metrics directly and plot here.
+def min_max_scale(input):
+ return (input - np.min(input)) / (np.max(input) - np.min(input))
+
+
+for i, sample in enumerate(test_data.test_dataloader()):
+ phase_image = sample["source"]
+ with torch.inference_mode(): # turn off gradient computation.
+ predicted_image = phase2fluor_model(phase_image)
+
+ target_image = (
+ sample["target"].cpu().numpy().squeeze(0)
+ ) # Squeezing batch dimension.
+ predicted_image = predicted_image.cpu().numpy().squeeze(0)
+ phase_image = phase_image.cpu().numpy().squeeze(0)
+ target_mem = min_max_scale(target_image[1, 0, :, :])
+ target_nuc = min_max_scale(target_image[0, 0, :, :])
+ # slicing channel dimension, squeezing z-dimension.
+ predicted_mem = min_max_scale(predicted_image[1, :, :, :].squeeze(0))
+ predicted_nuc = min_max_scale(predicted_image[0, :, :, :].squeeze(0))
+
+ # Compute SSIM and pearson correlation.
+ ssim_nuc = metrics.structural_similarity(target_nuc, predicted_nuc, data_range=1)
+ ssim_mem = metrics.structural_similarity(target_mem, predicted_mem, data_range=1)
+ pearson_nuc = np.corrcoef(target_nuc.flatten(), predicted_nuc.flatten())[0, 1]
+ pearson_mem = np.corrcoef(target_mem.flatten(), predicted_mem.flatten())[0, 1]
+
+ test_metrics.loc[i] = {
+ "pearson_nuc": pearson_nuc,
+ "SSIM_nuc": ssim_nuc,
+ "pearson_mem": pearson_mem,
+ "SSIM_mem": ssim_mem,
+ }
+
+test_metrics.boxplot(
+ column=["pearson_nuc", "SSIM_nuc", "pearson_mem", "SSIM_mem"],
+ rot=30,
+)
+# %%
+# Plot the predicted image
+channel_titles = ["Phase", "Nuclei", "Membrane"]
+fig, axes = plt.subplots(2, 3, figsize=(30, 20))
+
+for i, sample in enumerate(test_data.test_dataloader()):
+ # Plot the phase image
+ phase_image = sample["source"]
+ channel_image = phase_image[0, 0, 0]
+ p_low, p_high = np.percentile(channel_image, (0.5, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[0, 0].imshow(channel_image, cmap="gray")
+ axes[0, 0].axis("off")
+ axes[0, 0].set_title(channel_titles[0])
+
+ with torch.inference_mode(): # turn off gradient computation.
+ predicted_image = (
+ phase2fluor_model(phase_image.to(phase2fluor_model.device))
+ .cpu()
+ .numpy()
+ .squeeze(0)
+ )
+
+ target_image = sample["target"].cpu().numpy().squeeze(0)
+ # Plot the predicted images
+ for i in range(predicted_image.shape[-4]):
+ channel_image = predicted_image[i, 0]
+ p_low, p_high = np.percentile(channel_image, (0.1, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[0, i + 1].imshow(channel_image, cmap="gray")
+ axes[0, i + 1].axis("off")
+ axes[0, i + 1].set_title(f"VS {channel_titles[i + 1]}")
+
+ # Plot the target images
+ for i in range(target_image.shape[-4]):
+ channel_image = target_image[i, 0]
+ p_low, p_high = np.percentile(channel_image, (0.5, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[1, i].imshow(channel_image, cmap="gray")
+ axes[1, i].axis("off")
+ axes[1, i].set_title(f"Target {dataset.channel_names[i+1]}")
+
+ # Remove any unused subplots
+ for j in range(i + 1, 3):
+ fig.delaxes(axes[1, j])
+
+ plt.tight_layout()
+ plt.show()
+ break
+
+# %% [markdown] tags=[]
+"""
+
+
+
Task 2.2 Compute the metrics with respect to the pretrained model VSCyto2D
+Here we will compare your model with the VSCyto2D pretrained model by computing the pixel-based metrics and segmentation-based metrics.
+
+
+- When you ran the `setup.sh` you also downloaded the models in `/06_image_translation/pretrained_models/VSCyto2D/*.ckpt`
+- Load the VSCyto2 model model checkpoint and the configuration file
+- Compute the pixel-based metrics and segmentation-based metrics between the model you trained and the pretrained model
+
+
+
+We will evaluate the performance of your trained model with a pre-trained model using pixel based metrics as above and
+segmantation based metrics including (mAP@0.5, dice, accuracy and jaccard index).
+
+
+
Task 2.3 Compare the model's metrics
+In the previous section, we computed the pixel-based metrics and segmentation-based metrics.
+Now we will compare the performance of the model you trained with the pretrained model by plotting the boxplots.
+
+After you plot the metrics answer the following:
+
+- What do these metrics tells us about the performance of the model?
+- How do you interpret the differences in the metrics between the models?
+- How is your model compared to the pretrained model? How can you improve it?
+
+
+
+
+🎉 The end of the notebook 🎉
+Continue to Part 2: Image translation with generative models.
+
+
+Congratulations! You have trained several image translation models now!
+
+Please remember to document the hyperparameters,
+snapshots of predictions on validation set,
+and loss curves for your models and add the final performance in
+
+this google doc
+.
+We'll discuss our combined results as a group.
+