diff --git a/examples/demo_dlmbl/README.md b/examples/demo_dlmbl/README.md
index eb7917c8..b16378e5 100644
--- a/examples/demo_dlmbl/README.md
+++ b/examples/demo_dlmbl/README.md
@@ -1,6 +1,6 @@
-# Exercise 4: Image translation
+# Exercise 6: Image translation - Part 1
-This demo script was developed for the DL@MBL 2023 course by Ziwen Liu and Shalin Mehta, with many inputs and bugfixes by [Morgan Schwartz](https://github.com/msschwartz21), [Caroline Malin-Mayor](https://github.com/cmalinmayor), and [Peter Park](https://github.com/peterhpark).
+This demo script was developed for the DL@MBL 2024 course by Eduardo Hirata-Miyasaki, Ziwen Liu and Shalin Mehta, with many inputs and bugfixes by [Morgan Schwartz](https://github.com/msschwartz21), [Caroline Malin-Mayor](https://github.com/cmalinmayor), and [Peter Park](https://github.com/peterhpark).
@@ -9,10 +9,10 @@ This demo script was developed for the DL@MBL 2023 course by Ziwen Liu and Shali
Make sure that you are inside of the `image_translation` folder by using the `cd` command to change directories if needed.
-Make sure that you can use mamba to switch environments.
+Make sure that you can use conda to switch environments.
```bash
-mamba init
+conda init
```
**Close your shell, and login again.**
@@ -23,7 +23,7 @@ sh setup.sh
```
Activate your environment
```bash
-mamba activate 04_image_translation
+conda activate 06_image_translation
```
## Use vscode
@@ -42,7 +42,8 @@ jupyter notebook
...and continue with the instructions in the notebook.
-If 04_image_translation is not available as a kernel in jupyter, run
+If `06_image_translation` is not available as a kernel in jupyter, run:
+
```
-python -m ipykernel install --user --name=04_image_translation
+python -m ipykernel install --user --name=06_image_translation
```
diff --git a/examples/demo_dlmbl/convert-solution.py b/examples/demo_dlmbl/convert-solution.py
index 279f7874..91d7e46c 100644
--- a/examples/demo_dlmbl/convert-solution.py
+++ b/examples/demo_dlmbl/convert-solution.py
@@ -1,15 +1,15 @@
import argparse
-from traitlets.config import Config
-import nbformat as nbf
-from nbconvert.preprocessors import TagRemovePreprocessor, ClearOutputPreprocessor
+
from nbconvert.exporters import NotebookExporter
+from nbconvert.preprocessors import ClearOutputPreprocessor, TagRemovePreprocessor
+from traitlets.config import Config
def get_arg_parser():
parser = argparse.ArgumentParser()
- parser.add_argument('input_file')
- parser.add_argument('output_file')
+ parser.add_argument("input_file")
+ parser.add_argument("output_file")
return parser
@@ -21,7 +21,7 @@ def convert(input_file, output_file):
c.ClearOutputPreprocesser.enabled = True
c.NotebookExporter.preprocessors = [
"nbconvert.preprocessors.TagRemovePreprocessor",
- "nbconvert.preprocessors.ClearOutputPreprocessor"
+ "nbconvert.preprocessors.ClearOutputPreprocessor",
]
exporter = NotebookExporter(config=c)
@@ -29,7 +29,7 @@ def convert(input_file, output_file):
exporter.register_preprocessor(ClearOutputPreprocessor(), True)
output = NotebookExporter(config=c).from_filename(input_file)
- with open(output_file, 'w') as f:
+ with open(output_file, "w") as f:
f.write(output[0])
@@ -38,4 +38,4 @@ def convert(input_file, output_file):
args = parser.parse_args()
convert(args.input_file, args.output_file)
- print(f'Converted {args.input_file} to {args.output_file}')
+ print(f"Converted {args.input_file} to {args.output_file}")
diff --git a/examples/demo_dlmbl/debug_log_graph.py b/examples/demo_dlmbl/debug_log_graph.py
deleted file mode 100644
index ec987118..00000000
--- a/examples/demo_dlmbl/debug_log_graph.py
+++ /dev/null
@@ -1,97 +0,0 @@
-
-# %%
-# %% Imports and paths
-
-from pathlib import Path
-
-import matplotlib.pyplot as plt
-import numpy as np
-import pandas as pd
-import torch
-import torchview
-import torchvision
-from iohub import open_ome_zarr
-from lightning.pytorch import seed_everything
-from lightning.pytorch.loggers import CSVLogger
-
-# pytorch lightning wrapper for Tensorboard.
-from tensorboard import notebook # for viewing tensorboard in notebook
-from torch.utils.tensorboard import SummaryWriter # for logging to tensorboard
-
-# HCSDataModule makes it easy to load data during training.
-from viscy.data.hcs import HCSDataModule
-
-# Trainer class and UNet.
-from viscy.light.engine import VSUNet
-from viscy.light.trainer import VSTrainer
-
-seed_everything(42, workers=True)
-
-# Paths to data and log directory
-data_path = Path(
- Path("~/data/04_image_translation/HEK_nuclei_membrane_pyramid.zarr/")
-).expanduser()
-
-log_dir = Path("~/data/04_image_translation/logs/").expanduser()
-
-# Create log directory if needed, and launch tensorboard
-log_dir.mkdir(parents=True, exist_ok=True)
-
-# fmt: off
-%reload_ext tensorboard
-%tensorboard --logdir {log_dir} --port 6007 --bind_all
-# fmt: on
-
-# %% The entire training loop is contained in this cell.
-
-GPU_ID = 0
-BATCH_SIZE = 10
-YX_PATCH_SIZE = (512, 512)
-
-
-# Dictionary that specifies key parameters of the model.
-phase2fluor_config = {
- "architecture": "2D",
- "num_filters": [24, 48, 96, 192, 384],
- "in_channels": 1,
- "out_channels": 2,
- "residual": True,
- "dropout": 0.1, # dropout randomly turns off weights to avoid overfitting of the model to data.
- "task": "reg", # reg = regression task.
-}
-
-phase2fluor_model = VSUNet(
- model_config=phase2fluor_config.copy(),
- batch_size=BATCH_SIZE,
- loss_function=torch.nn.functional.l1_loss,
- schedule="WarmupCosine",
- log_num_samples=10, # Number of samples from each batch to log to tensorboard.
- example_input_yx_shape=YX_PATCH_SIZE,
-)
-
-# Reinitialize the data module.
-phase2fluor_data = HCSDataModule(
- data_path,
- source_channel="Phase",
- target_channel=["Nuclei", "Membrane"],
- z_window_size=1,
- split_ratio=0.8,
- batch_size=BATCH_SIZE,
- num_workers=8,
- architecture="2D",
- yx_patch_size=YX_PATCH_SIZE,
- augmentations=None,
-)
-phase2fluor_data.setup("fit")
-
-
-# Train for 3 epochs to see if you can log graph.
-trainer = VSTrainer(accelerator="gpu", devices=[GPU_ID], max_epochs=3, default_root_dir=log_dir)
-
-# trainer class takes the model and the data module as inputs.
-trainer.fit(phase2fluor_model, datamodule=phase2fluor_data)
-
-# %% Is exmple_input_array present?
-print(f'{phase2fluor_model.example_input_array.shape},{phase2fluor_model.example_input_array.dtype}')
-trainer.logger.log_graph(phase2fluor_model, phase2fluor_model.example_input_array)
-# %%
diff --git a/examples/demo_dlmbl/setup.sh b/examples/demo_dlmbl/setup.sh
index e502ceee..4b46f23f 100644
--- a/examples/demo_dlmbl/setup.sh
+++ b/examples/demo_dlmbl/setup.sh
@@ -2,31 +2,36 @@
START_DIR=$(pwd)
-# Create mamba environment
-mamba create -y --name 04_image_translation python=3.10
+# Create conda environment
+conda create -y --name 06_image_translation python=3.10
# Install ipykernel in the environment.
-mamba install -y ipykernel nbformat nbconvert black jupytext ipywidgets --name 04_image_translation
+conda install -y ipykernel nbformat nbconvert black jupytext ipywidgets --name 06_image_translation
# Specifying the environment explicitly.
-# mamba activate sometimes doesn't work from within shell scripts.
+# conda activate sometimes doesn't work from within shell scripts.
# install viscy and its dependencies`s in the environment using pip.
mkdir -p ~/code/
cd ~/code/
git clone https://github.com/mehta-lab/viscy.git
cd viscy
-git checkout 7c5e4c1d68e70163cf514d22c475da8ea7dc3a88 # Exercise is tested with this commit of viscy
-# Find path to the environment - mamba activate doesn't work from within shell scripts.
-ENV_PATH=$(conda info --envs | grep 04_image_translation | awk '{print $NF}')
+git checkout main #FIXME: change after merging this PR # Exercise is tested with this commit of viscy
+
+# Find path to the environment - conda activate doesn't work from within shell scripts.
+ENV_PATH=$(conda info --envs | grep 06_image_translation | awk '{print $NF}')
$ENV_PATH/bin/pip install ".[metrics]"
-# Create data directory
-mkdir -p ~/data/04_image_translation
-cd ~/data/04_image_translation
-wget https://dl-at-mbl-2023-data.s3.us-east-2.amazonaws.com/DLMBL2023_image_translation_data_pyramid.tar.gz
-wget https://dl-at-mbl-2023-data.s3.us-east-2.amazonaws.com/DLMBL2023_image_translation_test.tar.gz
-tar -xzf DLMBL2023_image_translation_data_pyramid.tar.gz
-tar -xzf DLMBL2023_image_translation_test.tar.gz
+# Create the directory structure
+mkdir -p ~/data/06_image_translation/training
+mkdir -p ~/data/06_image_translation/test
+
+# Change to the target directory
+cd ~/data/06_image_translation/training
+
+# Download the OME-Zarr dataset recursively
+wget -m -np -nH --cut-dirs=4 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VSCyto2D/training/a549_hoechst_cellmask_train_val.zarr/"
+cd ~/data/06_image_translation/test
+wget -m -np -nH --cut-dirs=4 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VSCyto2D/test/a549_hoechst_cellmask_test.zarr/"
# Change back to the starting directory
cd $START_DIR
diff --git a/examples/demo_dlmbl/solution.py b/examples/demo_dlmbl/solution.py
index 2c81aa6f..553fcf27 100644
--- a/examples/demo_dlmbl/solution.py
+++ b/examples/demo_dlmbl/solution.py
@@ -1,70 +1,115 @@
# %% [markdown]
"""
-# Image translation
----
+# Image translation (Virtual Staining)
-Written by Ziwen Liu and Shalin Mehta, CZ Biohub San Francisco.
+Written by Eduardo Hirata-Miyasaki, Ziwen Liu, and Shalin Mehta, CZ Biohub San Francisco.
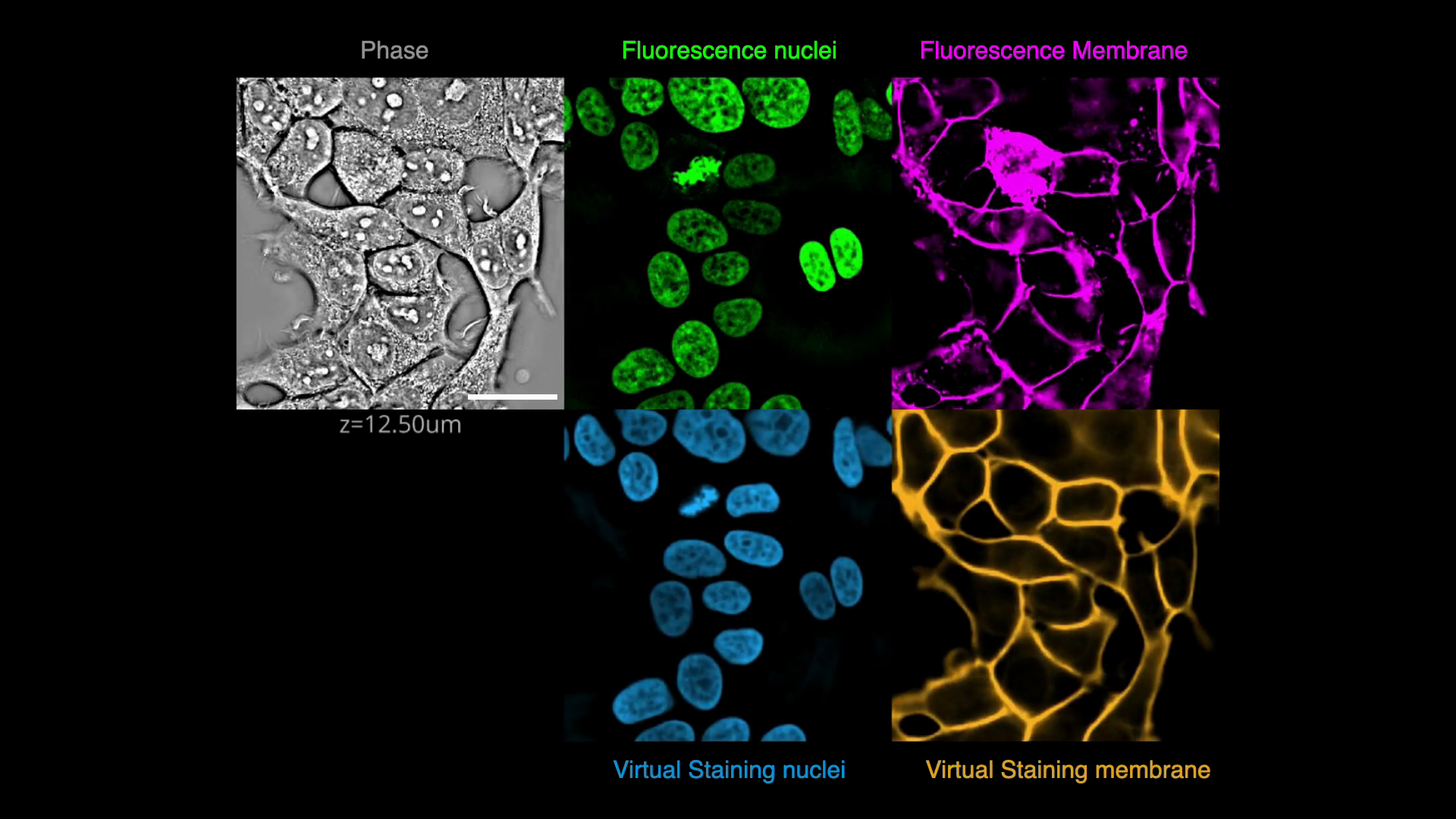
-In this exercise, we will solve an image translation task to predict fluorescence images of nuclei and membrane markers from quantitative phase images of cells. In other words, we will _virtually stain_ the nuclei and membrane visible in the phase image.
+## Overview
-Here, the source domain is label-free microscopy (material density) and the target domain is fluorescence microscopy (fluorophore density). The goal is to learn a mapping from the source domain to the target domain. We will use a deep convolutional neural network (CNN), specifically, a U-Net model with residual connections to learn the mapping. The preprocessing, training, prediction, evaluation, and deployment steps are unified in a computer vision pipeline for single-cell analysis that we call [VisCy](https://github.com/mehta-lab/VisCy).
+In this exercise, we will predict fluorescence images of
+nuclei and plasma membrane markers from quantitative phase images of cells,
+i.e., we will _virtually stain_ the nuclei and plasma membrane
+visible in the phase image.
+This is an example of an image translation task.
+We will apply spatial and intensity augmentations to train robust models
+and evaluate their performance.
+Finally, we will explore the opposite process of predicting a phase image
+from a fluorescence membrane label.
-VisCy evolved from our previous work on virtual staining of cellular components from their density and anisotropy.
-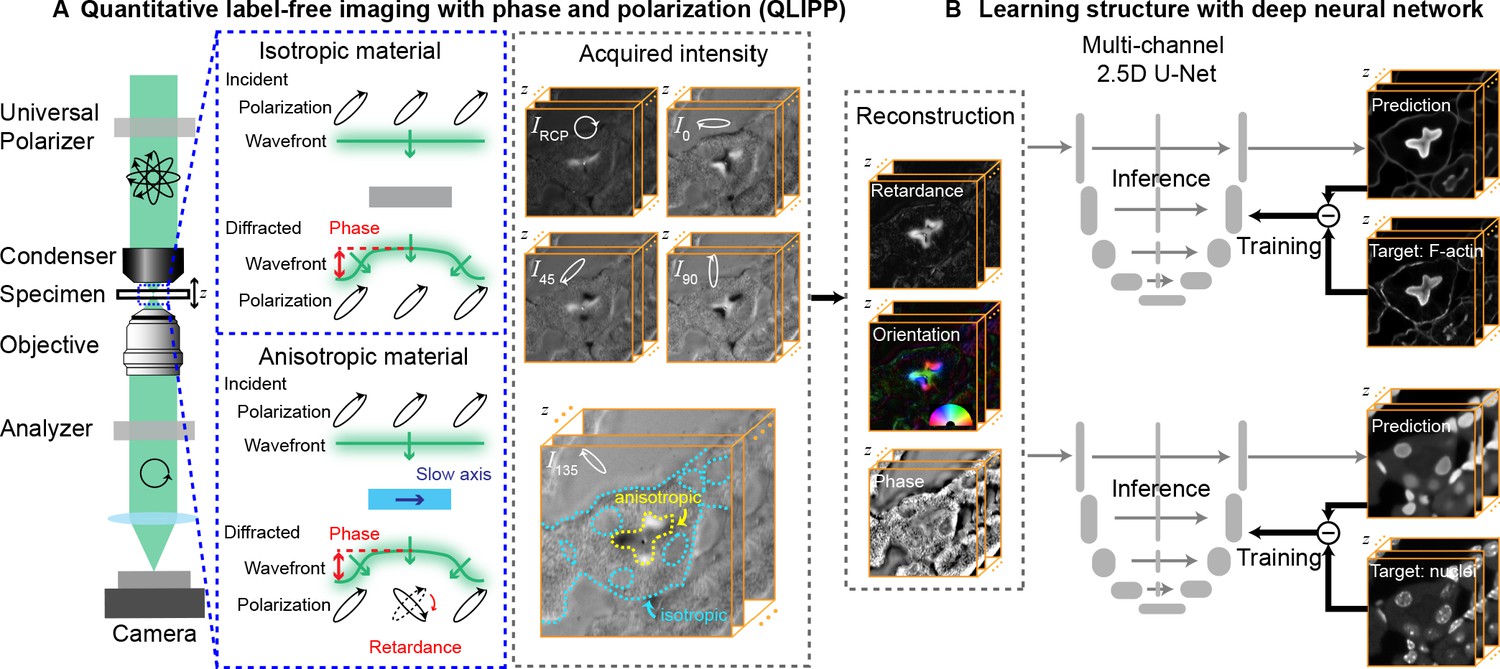
+[](https://github.com/mehta-lab/VisCy/assets/67518483/d53a81eb-eb37-44f3-b522-8bd7bddc7755)
+(Click on image to play video)
+"""
-[Guo et al. (2020) Revealing architectural order with quantitative label-free imaging and deep learning
-. eLife](https://elifesciences.org/articles/55502).
+# %% [markdown]
+"""
+### Goals
-VisCy exploits recent advances in the data and metadata formats ([OME-zarr](https://www.nature.com/articles/s41592-021-01326-w)) and DL frameworks, [PyTorch Lightning](https://lightning.ai/) and [MONAI](https://monai.io/).
+#### Part 1: Learn to use iohub (I/O library), VisCy dataloaders, and TensorBoard.
+
+ - Use a OME-Zarr dataset of 34 FOVs of adenocarcinomic human alveolar basal epithelial cells (A549),
+ each FOV has 3 channels (phase, nuclei, and cell membrane).
+ The nuclei were stained with DAPI and the cell membrane with Cellmask.
+ - Explore OME-Zarr using [iohub](https://czbiohub-sf.github.io/iohub/main/index.html)
+ and the high-content-screen (HCS) format.
+ - Use [MONAI](https://monai.io/) to implement data augmentations.
+
+#### Part 2: Train a model that predicts fluorescence from phase, and vice versa, using the UNeXt2 architecture.
+
+ - Create a model for image translation mapping from source domain to target domain
+ where the source domain is label-free microscopy (material density)
+ and the target domain is fluorescence microscopy (fluorophore density).
+ - Use the UNeXt2 architecture, a _purely convolutional architecture_
+ that draws on the design principles of transformer models to complete this task.
+ Here we will use a *UNeXt2*, an efficient image translation architecture inspired by ConvNeXt v2 and SparK.
+ - We will perform the preprocessing, training, prediction, evaluation, and deployment steps
+ that borrow from our computer vision pipeline for single-cell analysis in
+ our pipeline called [VisCy](https://github.com/mehta-lab/VisCy).
+ - Reuse the same architecture as above and create a similar model doing the inverse task (fluorescence to phase).
+ - Evaluate the model.
+
+#### (Extra) Play with the hyperparameters to improve the models or train a 3D UNeXt2
+
+Our guesstimate is that each of the three parts will take ~1-1.5 hours.
+A reasonable 2D UNet can be trained in ~30 min on a typical AWS node.
+The focus of the exercise is on understanding the information content of the data,
+how to train and evaluate 2D image translation models, and exploring some hyperparameters of the model.
+If you complete this exercise and have time to spare, try the bonus exercise on 3D image translation.
+
+Checkout [VisCy](https://github.com/mehta-lab/VisCy/tree/main/examples/demos),
+our deep learning pipeline for training and deploying computer vision models
+for image-based phenotyping including the robust virtual staining of landmark organelles.
+VisCy exploits recent advances in data and metadata formats
+([OME-zarr](https://www.nature.com/articles/s41592-021-01326-w)) and DL frameworks,
+[PyTorch Lightning](https://lightning.ai/) and [MONAI](https://monai.io/).
+
+### References
+
+- [Liu, Z. and Hirata-Miyasaki, E. et al. (2024) Robust Virtual Staining of Cellular Landmarks](https://www.biorxiv.org/content/10.1101/2024.05.31.596901v2.full.pdf)
+- [Guo et al. (2020) Revealing architectural order with quantitative label-free imaging and deep learning. eLife](https://elifesciences.org/articles/55502)
"""
+
# %% [markdown]
"""
-Today, we will train a 2D image translation model using a 2D U-Net with residual connections. We will use a dataset of 301 fields of view (FOVs) of Human Embryonic Kidney (HEK) cells, each FOV has 3 channels (phase, membrane, and nuclei). The cells were labeled with CRISPR editing. Intrestingly, not all cells during this experiment were labeled due to the stochastic nature of CRISPR editing. In such situations, virtual staining rescues missing labels.
-
+📖 As you work through parts 2 and 3, please share the layouts of your models (output of torchview)
+and their performance with everyone via
+[this Google Doc](https://docs.google.com/document/d/1Mq-yV8FTG02xE46Mii2vzPJVYSRNdeOXkeU-EKu-irE/edit?usp=sharing). 📖
"""
# %% [markdown]
"""
-The exercise is organized in 3 parts.
+The exercise is organized in 3 parts + Extra part.
+
+
+- Part 1 - Learn to use iohub (I/O library), VisCy dataloaders, and tensorboard.
+- Part 2 - Train and evaluate the model to translate phase into fluorescence, and vice versa.
+- Extra task - Tune the models to improve performance.
+
-* **Part 1** - Explore the data using tensorboard. Launch the training before lunch.
-* Lunch break - The model will continue training during lunch.
-* **Part 2** - Evaluate the training with tensorboard. Train another model.
-* **Part 3** - Tune the models to improve performance.
-Set your python kernel to 04_image_translation
+Set your python kernel to 06_image_translation
"""
-# %%
+# %% [markdown]
"""
-# Part 1: Log training data to tensorboard, start training a model.
+## Part 1: Log training data to tensorboard, start training a model.
---------
-
Learning goals:
-- Load the OME-zarr dataset and examine the channels.
+- Load the OME-zarr dataset and examine the channels (A549).
- Configure and understand the data loader.
- Log some patches to tensorboard.
-- Initialize a 2D U-Net model for virtual staining
+- Initialize a 2D UNeXt2 model for virtual staining of nuclei and membrane from phase.
- Start training the model to predict nuclei and membrane from phase.
"""
-# %% Imports and paths
+# %% Imports
+import os
from pathlib import Path
import matplotlib.pyplot as plt
@@ -74,19 +119,24 @@
import torchview
import torchvision
from iohub import open_ome_zarr
+from iohub.reader import print_info
from lightning.pytorch import seed_everything
-from lightning.pytorch.loggers import CSVLogger, TensorBoardLogger
+from lightning.pytorch.loggers import TensorBoardLogger
from skimage import metrics # for metrics.
-# %% Imports and paths
# pytorch lightning wrapper for Tensorboard.
from torch.utils.tensorboard import SummaryWriter # for logging to tensorboard
# HCSDataModule makes it easy to load data during training.
from viscy.data.hcs import HCSDataModule
+# Trainer class and UNet.
+from viscy.light.engine import MixedLoss, VSUNet
+from viscy.light.trainer import VSTrainer
+
# training augmentations
from viscy.transforms import (
+ NormalizeSampled,
RandAdjustContrastd,
RandAffined,
RandGaussianNoised,
@@ -95,62 +145,115 @@
RandWeightedCropd,
)
-# Trainer class and UNet.
-from viscy.light.engine import VSUNet
-from viscy.light.trainer import VSTrainer
-
+# %%
+# seed random number generators for reproducibility.
seed_everything(42, workers=True)
# Paths to data and log directory
-data_path = Path(
- Path("~/data/04_image_translation/HEK_nuclei_membrane_pyramid.zarr/")
-).expanduser()
+top_dir = Path(
+ f"/hpc/mydata/{os.environ['USER']}/data/"
+) # TODO: Change this to point to your data directory.
+data_path = (
+ top_dir / "06_image_translation/training/a549_hoechst_cellmask_train_val.zarr"
+)
+log_dir = top_dir / "06_image_translation/logs/"
-log_dir = Path("~/data/04_image_translation/logs/").expanduser()
+if not data_path.exists():
+ raise FileNotFoundError(
+ f"Data not found at {data_path}. Please check the top_dir and data_path variables."
+ )
+# %%
# Create log directory if needed, and launch tensorboard
log_dir.mkdir(parents=True, exist_ok=True)
# %% [markdown] tags=[]
"""
-The next cell starts tensorboard within the notebook.
+The next cell starts tensorboard.
-
+
If you launched jupyter lab from ssh terminal, add --host <your-server-name> to the tensorboard command below. <your-server-name> is the address of your compute node that ends in amazonaws.com.
-You can also launch tensorboard in an independent tab (instead of in the notebook) by changing the `%` to `!`
+
+
+
+If you are using VSCode and a remote server, you will need to forward the port to view the tensorboard.
+Take note of the port number was assigned in the previous cell.(i.e
http://localhost:{port_number_assigned})
+
+Locate the your VSCode terminal and select the
Ports tab
+
+- Add a new port with the
port_number_assigned
+
+Click on the link to view the tensorboard and it should open in your browser.
"""
# %% Imports and paths tags=[]
-%reload_ext tensorboard
-%tensorboard --logdir {log_dir}
+
+
+# Function to find an available port
+def find_free_port():
+ import socket
+
+ with socket.socket(socket.AF_INET, socket.SOCK_STREAM) as s:
+ s.bind(("", 0))
+ return s.getsockname()[1]
+
+
+# Launch TensorBoard on the browser
+def launch_tensorboard(log_dir):
+ import subprocess
+
+ port = find_free_port()
+ tensorboard_cmd = f"tensorboard --logdir={log_dir} --port={port}"
+ process = subprocess.Popen(tensorboard_cmd, shell=True)
+ print(
+ f"TensorBoard started at http://localhost:{port}. \n"
+ "If you are using VSCode remote session, forward the port using the PORTS tab next to TERMINAL."
+ )
+ return process
+
+
+# Launch tensorboard and click on the link to view the logs.
+tensorboard_process = launch_tensorboard(log_dir)
# %% [markdown]
"""
-## Load Dataset.
+## Load OME-Zarr Dataset
-There should be 301 FOVs in the dataset (12 GB compressed).
+There should be 34 FOVs in the dataset.
Each FOV consists of 3 channels of 2048x2048 images,
-saved in the
-High-Content Screening (HCS) layout
+saved in the [High-Content Screening (HCS) layout](https://ngff.openmicroscopy.org/latest/#hcs-layout)
specified by the Open Microscopy Environment Next Generation File Format
(OME-NGFF).
-The layout on the disk is: row/col/field/pyramid_level/timepoint/channel/z/y/x.
-Notice that labelling of nuclei channel is not complete - some cells are not expressing the fluorescent protein.
+- The layout on the disk is: `row/col/field/pyramid_level/timepoint/channel/z/y/x.`
"""
+# %% [markdown]
+"""
+
+You can inspect the tree structure by using your terminal:
+ iohub info -v "path-to-ome-zarr"
+
+
+More info on the CLI:
+iohub info --help to see the help menu.
+
+"""
# %%
-dataset = open_ome_zarr(data_path)
+# This is the python function called by `iohub info` CLI command
+print_info(data_path, verbose=True)
-print(f"Number of positions: {len(list(dataset.positions()))}")
+# Open and inspect the dataset.
+dataset = open_ome_zarr(data_path)
+# %%
# Use the field and pyramid_level below to visualize data.
row = 0
col = 0
-field = 23 # TODO: Change this to explore data.
+field = 9 # TODO: Change this to explore data.
# This dataset contains images at 3 resolutions.
# '0' is the highest resolution
@@ -183,7 +286,8 @@
#
# ### Task 1.1
#
-# Look at a couple different fields of view by changing the value in the cell above. See if you notice any missing or inconsistent staining.
+# Look at a couple different fields of view by changing the value in the cell above.
+# Check the cell density, the cell morphologies, and fluorescence signal.
#
+#
+# ### Task 1.3
+# Add augmentations to the datamodule and rerun the setup.
+#
+# What kind of augmentations do you think are important for this task?
+#
+# How do they make the model more robust?
+#
+# Add augmentations to rotate about $\pi$ around z-axis, 30% scale in y,x,
+# shearing of 10% and no padding with zeros with a probablity of 80%.
+#
+# Add a Gaussian noise with a mean of 0.0 and standard deviation of 0.3 with a probability of 50%.
+#
+# HINT: `RandAffined()` and `RandGaussianNoised()` are from
+# `viscy.transforms` [here](https://github.com/mehta-lab/VisCy/blob/main/viscy/transforms.py).
+# *Note these are MONAI transforms that have been redefined for VisCy.*
+# Can you tell what augmentation were applied from looking at the augmented images in Tensorboard?
+#
+# HINT:
+# [Compare your choice of augmentations by dowloading the pretrained models and config files](https://github.com/mehta-lab/VisCy/releases/download/v0.1.0/VisCy-0.1.0-VS-models.zip).
+#
# %%
# Here we turn on data augmentation and rerun setup
+source_channel = ["Phase3D"]
+target_channel = ["Nucl", "Mem"]
+
augmentations = [
RandWeightedCropd(
- keys=["Phase", "Membrane", "Nuclei"], w_key="Nuclei", spatial_size=[512, 512]
+ keys=source_channel + target_channel,
+ spatial_size=(1, 256, 256),
+ num_samples=2,
+ w_key=target_channel[0],
),
- RandAffined(
- keys=["Phase", "Membrane", "Nuclei"],
+ RandAdjustContrastd(keys=source_channel, prob=0.5, gamma=(0.8, 1.2)),
+ RandScaleIntensityd(keys=source_channel, factors=0.5, prob=0.5),
+ RandGaussianSmoothd(
+ keys=source_channel,
+ sigma_x=(0.25, 0.75),
+ sigma_y=(0.25, 0.75),
+ sigma_z=(0.0, 0.0),
prob=0.5,
+ ),
+ # #######################
+ # ##### TODO ########
+ # #######################
+ ##TODO: Add rotation agumentations
+ ## Write code below
+ ## TODO: Add Random Gaussian Noise
+ ## Write code below
+]
+
+normalizations = [
+ NormalizeSampled(
+ keys=source_channel + target_channel,
+ level="fov_statistics",
+ subtrahend="mean",
+ divisor="std",
+ )
+]
+
+data_module.augmentations = augmentations
+data_module.setup("fit")
+
+# get the new data loader with augmentation turned on
+augmented_train_dataloader = data_module.train_dataloader()
+
+# Draw batches and write to tensorboard
+writer = SummaryWriter(log_dir=f"{log_dir}/view_batch")
+augmented_batch = next(iter(augmented_train_dataloader))
+log_batch_tensorboard(augmented_batch, 0, writer, "augmentation/some")
+writer.close()
+
+# %% tags=["solution"]
+# #######################
+# ##### SOLUTION ########
+# #######################
+source_channel = ["Phase3D"]
+target_channel = ["Nucl", "Mem"]
+
+augmentations = [
+ RandWeightedCropd(
+ keys=source_channel + target_channel,
+ spatial_size=(1, 384, 384),
+ num_samples=2,
+ w_key=target_channel[0],
+ ),
+ RandAffined(
+ keys=source_channel + target_channel,
rotate_range=[3.14, 0.0, 0.0],
- shear_range=[0.0, 0.05, 0.05],
scale_range=[0.0, 0.3, 0.3],
+ prob=0.8,
+ padding_mode="zeros",
+ shear_range=[0.0, 0.01, 0.01],
),
- RandAdjustContrastd(keys=["Phase"], prob=0.3, gamma=[0.5, 1.5]),
- RandScaleIntensityd(keys=["Phase"], prob=0.5, factors=0.5),
- RandGaussianNoised(keys=["Phase"], prob=0.5, std=1),
+ RandAdjustContrastd(keys=source_channel, prob=0.5, gamma=(0.8, 1.2)),
+ RandScaleIntensityd(keys=source_channel, factors=0.5, prob=0.5),
+ RandGaussianNoised(keys=source_channel, prob=0.5, mean=0.0, std=0.3),
RandGaussianSmoothd(
- keys=["Phase"], prob=0.5, sigma_x=[0.25, 1.5], sigma_y=[0.25, 1.5]
+ keys=source_channel,
+ sigma_x=(0.25, 0.75),
+ sigma_y=(0.25, 0.75),
+ sigma_z=(0.0, 0.0),
+ prob=0.5,
),
]
+normalizations = [
+ NormalizeSampled(
+ keys=source_channel + target_channel,
+ level="fov_statistics",
+ subtrahend="mean",
+ divisor="std",
+ )
+]
+
data_module.augmentations = augmentations
data_module.setup("fit")
@@ -383,119 +580,129 @@ def log_batch_jupyter(batch):
log_batch_tensorboard(augmented_batch, 0, writer, "augmentation/some")
writer.close()
+
# %% [markdown]
-# Visualize directly on Jupyter ☄️
+# Visualize directly on Jupyter
# %%
log_batch_jupyter(augmented_batch)
-# %% [markdown]
-#
-#
-# ### Task 1.3
-# Can you tell what augmentation were applied from looking at the augmented images in Tensorboard?
-#
-# Check your answer using the source code [here](https://github.com/mehta-lab/VisCy/blob/b89f778b34735553cf155904eef134c756708ff2/viscy/light/data.py#L529).
-#
-
# %% [markdown]
"""
## Train a 2D U-Net model to predict nuclei and membrane from phase.
-### Construct a 2D U-Net
+### Construct a 2D UNeXt2 using VisCy
See ``viscy.unet.networks.Unet2D.Unet2d`` ([source code](https://github.com/mehta-lab/VisCy/blob/7c5e4c1d68e70163cf514d22c475da8ea7dc3a88/viscy/unet/networks/Unet2D.py#L7)) for configuration details.
"""
# %%
# Create a 2D UNet.
GPU_ID = 0
-BATCH_SIZE = 10
-YX_PATCH_SIZE = (512, 512)
+BATCH_SIZE = 12
+YX_PATCH_SIZE = (256, 256)
# Dictionary that specifies key parameters of the model.
-phase2fluor_config = {
- "architecture": "2D",
- "num_filters": [24, 48, 96, 192, 384],
- "in_channels": 1,
- "out_channels": 2,
- "residual": True,
- "dropout": 0.1, # dropout randomly turns off weights to avoid overfitting of the model to data.
- "task": "reg", # reg = regression task.
-}
+
+phase2fluor_config = dict(
+ in_channels=1,
+ out_channels=2,
+ encoder_blocks=[3, 3, 9, 3],
+ dims=[96, 192, 384, 768],
+ decoder_conv_blocks=2,
+ stem_kernel_size=(1, 2, 2),
+ in_stack_depth=1,
+ pretraining=False,
+)
phase2fluor_model = VSUNet(
+ architecture="UNeXt2_2D", # 2D UNeXt2 architecture
model_config=phase2fluor_config.copy(),
- batch_size=BATCH_SIZE,
- loss_function=torch.nn.functional.l1_loss,
+ loss_function=MixedLoss(l1_alpha=0.5, l2_alpha=0.0, ms_dssim_alpha=0.5),
schedule="WarmupCosine",
- log_num_samples=5, # Number of samples from each batch to log to tensorboard.
- example_input_yx_shape=YX_PATCH_SIZE,
+ lr=2e-4,
+ log_batches_per_epoch=5, # Number of samples from each batch to log to tensorboard.
+ freeze_encoder=False,
)
-
# %% [markdown]
"""
### Instantiate data module and trainer, test that we are setup to launch training.
"""
# %%
+source_channel = ["Phase3D"]
+target_channel = ["Nucl", "Mem"]
# Setup the data module.
-phase2fluor_data = HCSDataModule(
+phase2fluor_2D_data = HCSDataModule(
data_path,
- source_channel="Phase",
- target_channel=["Membrane", "Nuclei"],
+ architecture="UNeXt2_2D",
+ source_channel=source_channel,
+ target_channel=target_channel,
z_window_size=1,
split_ratio=0.8,
batch_size=BATCH_SIZE,
num_workers=8,
- architecture="2D",
yx_patch_size=YX_PATCH_SIZE,
augmentations=augmentations,
+ normalizations=normalizations,
)
-phase2fluor_data.setup("fit")
+phase2fluor_2D_data.setup("fit")
# fast_dev_run runs a single batch of data through the model to check for errors.
trainer = VSTrainer(accelerator="gpu", devices=[GPU_ID], fast_dev_run=True)
# trainer class takes the model and the data module as inputs.
-trainer.fit(phase2fluor_model, datamodule=phase2fluor_data)
+trainer.fit(phase2fluor_model, datamodule=phase2fluor_2D_data)
# %% [markdown]
# ## View model graph.
#
-# PyTorch uses dynamic graphs under the hood. The graphs are constructed on the fly. This is in contrast to TensorFlow, where the graph is constructed before the training loop and remains static. In other words, the graph of the network can change with every forward pass. Therefore, we need to supply an input tensor to construct the graph. The input tensor can be a random tensor of the correct shape and type. We can also supply a real image from the dataset. The latter is more useful for debugging.
+# PyTorch uses dynamic graphs under the hood.
+# The graphs are constructed on the fly.
+# This is in contrast to TensorFlow,
+# where the graph is constructed before the training loop and remains static.
+# In other words, the graph of the network can change with every forward pass.
+# Therefore, we need to supply an input tensor to construct the graph.
+# The input tensor can be a random tensor of the correct shape and type.
+# We can also supply a real image from the dataset.
+# The latter is more useful for debugging.
# %% [markdown]
#
#
# ### Task 1.4
-# Run the next cell to generate a graph representation of the model architecture. Can you recognize the UNet structure and skip connections in this graph visualization?
+# Run the next cell to generate a graph representation of the model architecture.
+# Can you recognize the UNet structure and skip connections in this graph visualization?
#
# %%
# visualize graph of phase2fluor model as image.
model_graph_phase2fluor = torchview.draw_graph(
phase2fluor_model,
- phase2fluor_data.train_dataset[0]["source"],
- depth=2, # adjust depth to zoom in.
+ phase2fluor_2D_data.train_dataset[0]["source"][0].unsqueeze(dim=0),
+ roll=True,
+ depth=3, # adjust depth to zoom in.
device="cpu",
+ # expand_nested=True,
)
# Print the image of the model.
model_graph_phase2fluor.visual_graph
+
# %% [markdown]
"""
-### Task 1.5
+
Task 1.5
Start training by running the following cell. Check the new logs on the tensorboard.
"""
-
# %%
-
+# Check if GPU is available
+# You can check by typing `nvidia-smi`
GPU_ID = 0
-n_samples = len(phase2fluor_data.train_dataset)
+
+n_samples = len(phase2fluor_2D_data.train_dataset)
steps_per_epoch = n_samples // BATCH_SIZE # steps per epoch.
n_epochs = 50 # Set this to 50 or the number of epochs you want to train for.
@@ -513,41 +720,43 @@ def log_batch_jupyter(batch):
),
)
# Launch training and check that loss and images are being logged on tensorboard.
-trainer.fit(phase2fluor_model, datamodule=phase2fluor_data)
+trainer.fit(phase2fluor_model, datamodule=phase2fluor_2D_data)
# %% [markdown]
"""
-## Checkpoint 1
+
Checkpoint 1
Now the training has started,
we can come back after a while and evaluate the performance!
-
-"""
-# %%
-"""
-# Part 2: Assess previous model, train fluorescence to phase contrast translation model.
---------------------------------------------------
+
"""
# %% [markdown]
"""
-We now look at some metrics of performance of previous model. We typically evaluate the model performance on a held out test data. We will use the following metrics to evaluate the accuracy of regression of the model:
+## Part 2: Assess previous model, train fluorescence to phase contrast translation model.
+
+We now look at some metrics of performance of previous model.
+We typically evaluate the model performance on a held out test data.
+We will use the following metrics to evaluate the accuracy of regression of the model:
+
- [Person Correlation](https://en.wikipedia.org/wiki/Pearson_correlation_coefficient).
- [Structural similarity](https://en.wikipedia.org/wiki/Structural_similarity) (SSIM).
-You should also look at the validation samples on tensorboard (hint: the experimental data in nuclei channel is imperfect.)
+You should also look at the validation samples on tensorboard
+(hint: the experimental data in nuclei channel is imperfect.)
"""
# %% [markdown]
"""
-### Task 2.1 Define metrics
+
Task 2.1 Define metrics
-For each of the above metrics, write a brief definition of what they are and what they mean for this image translation task.
+For each of the above metrics, write a brief definition of what they are and what they mean
+for this image translation task. Use your favorite search engine and/or resources.
"""
@@ -557,25 +766,31 @@ def log_batch_jupyter(batch):
# #######################
# ##### Todo ############
# #######################
+#
# ```
#
# - Pearson Correlation:
#
# - Structural similarity:
-# %% Compute metrics directly and plot here.
-test_data_path = Path(
- "~/data/04_image_translation/HEK_nuclei_membrane_test.zarr"
-).expanduser()
+# %% [markdown]
+"""
+Let's compute metrics directly and plot below.
+"""
+# %%
+# Setup the test data module.
+test_data_path = top_dir / "06_image_translation/test/a549_hoechst_cellmask_test.zarr"
+source_channel = ["Phase3D"]
+target_channel = ["Nucl", "Mem"]
test_data = HCSDataModule(
test_data_path,
- source_channel="Phase",
- target_channel=["Membrane", "Nuclei"],
+ source_channel=source_channel,
+ target_channel=target_channel,
z_window_size=1,
batch_size=1,
num_workers=8,
- architecture="2D",
+ architecture="UNeXt2",
)
test_data.setup("test")
@@ -583,12 +798,13 @@ def log_batch_jupyter(batch):
columns=["pearson_nuc", "SSIM_nuc", "pearson_mem", "SSIM_mem"]
)
+# %% Compute metrics directly and plot here.
+
def min_max_scale(input):
return (input - np.min(input)) / (np.max(input) - np.min(input))
-# %% Compute metrics directly and plot here.
for i, sample in enumerate(test_data.test_dataloader()):
phase_image = sample["source"]
with torch.inference_mode(): # turn off gradient computation.
@@ -622,13 +838,60 @@ def min_max_scale(input):
column=["pearson_nuc", "SSIM_nuc", "pearson_mem", "SSIM_mem"],
rot=30,
)
+# %%
+# Plot the predicted image
+channel_titles = ["Phase", "Nuclei", "Membrane"]
+fig, axes = plt.subplots(2, 3, figsize=(30, 20))
+for i, sample in enumerate(test_data.test_dataloader()):
+ # Plot the phase image
+ phase_image = sample["source"]
+ channel_image = phase_image[0, 0, 0]
+ p_low, p_high = np.percentile(channel_image, (0.5, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[0, 0].imshow(channel_image, cmap="gray")
+ axes[0, 0].axis("off")
+ axes[0, 0].set_title(channel_titles[0])
+
+ with torch.inference_mode(): # turn off gradient computation.
+ predicted_image = (
+ phase2fluor_model(phase_image.to(phase2fluor_model.device))
+ .cpu()
+ .numpy()
+ .squeeze(0)
+ )
+
+ target_image = sample["target"].cpu().numpy().squeeze(0)
+ # Plot the predicted images
+ for i in range(predicted_image.shape[-4]):
+ channel_image = predicted_image[i, 0]
+ p_low, p_high = np.percentile(channel_image, (0.1, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[0, i + 1].imshow(channel_image, cmap="gray")
+ axes[0, i + 1].axis("off")
+ axes[0, i + 1].set_title(f"VS {channel_titles[i + 1]}")
+ # Plot the target images
+ for i in range(target_image.shape[-4]):
+ channel_image = target_image[i, 0]
+ p_low, p_high = np.percentile(channel_image, (0.5, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[1, i].imshow(channel_image, cmap="gray")
+ axes[1, i].axis("off")
+ axes[1, i].set_title(f"Target {dataset.channel_names[i+1]}")
+
+ # Remove any unused subplots
+ for j in range(i + 1, 3):
+ fig.delaxes(axes[1, j])
+
+ plt.tight_layout()
+ plt.show()
+ break
# %% [markdown] tags=[]
"""
-### Task 2.2 Train fluorescence to phase contrast translation model
+
Task 2.2 Train fluorescence to phase contrast translation model
Instantiate a data module, model, and trainer for fluorescence to phase contrast translation. Copy over the code from previous cells and update the parameters. Give the variables and paths a different name/suffix (fluor2phase) to avoid overwriting objects used to train phase2fluor models.
@@ -652,12 +915,6 @@ def min_max_scale(input):
# Your code here (copy from above and modify as needed)
)
-trainer = VSTrainer(
- # Your code here (copy from above and modify as needed)
-)
-trainer.fit(fluor2phase_model, datamodule=fluor2phase_data)
-
-
# Visualize the graph of fluor2phase model as image.
model_graph_fluor2phase = torchview.draw_graph(
fluor2phase_model,
@@ -674,42 +931,112 @@ def min_max_scale(input):
##########################
# The entire training loop is contained in this cell.
+source_channel = ["Mem"] # or 'Nuc' depending on choice
+target_channel = ["Phase3D"]
+YX_PATCH_SIZE = (256, 256)
+BATCH_SIZE = 12
+n_epochs = 50
+
+# Setup the new augmentations
+augmentations = [
+ RandWeightedCropd(
+ keys=source_channel + target_channel,
+ spatial_size=(1, 384, 384),
+ num_samples=2,
+ w_key=target_channel[0],
+ ),
+ RandAffined(
+ keys=source_channel + target_channel,
+ rotate_range=[3.14, 0.0, 0.0],
+ scale_range=[0.0, 0.3, 0.3],
+ prob=0.8,
+ padding_mode="zeros",
+ shear_range=[0.0, 0.01, 0.01],
+ ),
+ RandAdjustContrastd(keys=source_channel, prob=0.5, gamma=(0.8, 1.2)),
+ RandScaleIntensityd(keys=source_channel, factors=0.5, prob=0.5),
+ RandGaussianNoised(keys=source_channel, prob=0.5, mean=0.0, std=0.3),
+ RandGaussianSmoothd(
+ keys=source_channel,
+ sigma_x=(0.25, 0.75),
+ sigma_y=(0.25, 0.75),
+ sigma_z=(0.0, 0.0),
+ prob=0.5,
+ ),
+]
+
+normalizations = [
+ NormalizeSampled(
+ keys=source_channel + target_channel,
+ level="fov_statistics",
+ subtrahend="mean",
+ divisor="std",
+ )
+]
+# Setup the dataloader
fluor2phase_data = HCSDataModule(
data_path,
- source_channel="Membrane",
- target_channel="Phase",
+ architecture="UNeXt2_2D",
+ source_channel=source_channel,
+ target_channel=target_channel,
z_window_size=1,
split_ratio=0.8,
batch_size=BATCH_SIZE,
num_workers=8,
- architecture="2D",
yx_patch_size=YX_PATCH_SIZE,
augmentations=augmentations,
+ normalizations=normalizations,
)
fluor2phase_data.setup("fit")
+n_samples = len(fluor2phase_data.train_dataset)
+
+steps_per_epoch = n_samples // BATCH_SIZE # steps per epoch.
+
# Dictionary that specifies key parameters of the model.
-fluor2phase_config = {
- "architecture": "2D",
- "in_channels": 1,
- "out_channels": 1,
- "residual": True,
- "dropout": 0.1, # dropout randomly turns off weights to avoid overfitting of the model to data.
- "task": "reg", # reg = regression task.
- "num_filters": [24, 48, 96, 192, 384],
-}
+fluor2phase_config = dict(
+ in_channels=1,
+ out_channels=1,
+ encoder_blocks=[3, 3, 9, 3],
+ dims=[96, 192, 384, 768],
+ decoder_conv_blocks=2,
+ stem_kernel_size=(1, 2, 2),
+ in_stack_depth=1,
+ pretraining=False,
+)
fluor2phase_model = VSUNet(
+ architecture="UNeXt2_2D",
model_config=fluor2phase_config.copy(),
- batch_size=BATCH_SIZE,
- loss_function=torch.nn.functional.mse_loss,
+ loss_function=MixedLoss(l1_alpha=0.5, l2_alpha=0.0, ms_dssim_alpha=0.5),
schedule="WarmupCosine",
- log_num_samples=5,
- example_input_yx_shape=YX_PATCH_SIZE,
+ lr=2e-4,
+ log_batches_per_epoch=5, # Number of samples from each batch to log to tensorboard.
+ freeze_encoder=False,
+)
+
+# Visualize the graph of fluor2phase model as image.
+model_graph_fluor2phase = torchview.draw_graph(
+ fluor2phase_model,
+ next(iter(fluor2phase_data.train_dataloader()))["source"],
+ depth=3, # adjust depth to zoom in.
+ device="cpu",
+)
+model_graph_fluor2phase.visual_graph
+
+# %% tags=[]
+##########################
+######## TODO ########
+##########################
+
+trainer = VSTrainer(
+ # Your code here (copy from above and modify as needed)
)
+trainer.fit(fluor2phase_model, datamodule=fluor2phase_data)
+# %% tags=["solution"]
trainer = VSTrainer(
accelerator="gpu",
devices=[GPU_ID],
@@ -725,20 +1052,11 @@ def min_max_scale(input):
trainer.fit(fluor2phase_model, datamodule=fluor2phase_data)
-# Visualize the graph of fluor2phase model as image.
-model_graph_fluor2phase = torchview.draw_graph(
- fluor2phase_model,
- fluor2phase_data.train_dataset[0]["source"],
- depth=2, # adjust depth to zoom in.
- device="cpu",
-)
-model_graph_fluor2phase.visual_graph
-
# %% [markdown] tags=[]
"""
-### Task 2.3
+
Task 2.3
While your model is training, let's think about the following questions:
- What is the information content of each channel in the dataset?
@@ -748,32 +1066,28 @@ def min_max_scale(input):
"""
# %%
test_data_path = Path(
- "~/data/04_image_translation/HEK_nuclei_membrane_test.zarr"
+ "~/data/06_image_translation/test/a549_hoechst_cellmask_test.zarr"
).expanduser()
test_data = HCSDataModule(
test_data_path,
- source_channel="Nuclei", # or Membrane, depending on your choice of source
- target_channel="Phase",
+ source_channel="Mem", # or Nuc, depending on your choice of source
+ target_channel="Phase3D",
z_window_size=1,
batch_size=1,
num_workers=8,
- architecture="2D",
+ architecture="UNeXt2",
)
test_data.setup("test")
test_metrics = pd.DataFrame(columns=["pearson_phase", "SSIM_phase"])
-def min_max_scale(input):
- return (input - np.min(input)) / (np.max(input) - np.min(input))
-
-
# %%
for i, sample in enumerate(test_data.test_dataloader()):
source_image = sample["source"]
with torch.inference_mode(): # turn off gradient computation.
- predicted_image = fluor2phase_model(source_image)
+ predicted_image = fluor2phase_model(source_image.to(fluor2phase_model.device))
target_image = (
sample["target"].cpu().numpy().squeeze(0)
@@ -799,42 +1113,96 @@ def min_max_scale(input):
column=["pearson_phase", "SSIM_phase"],
rot=30,
)
+# %%
+# Plot the predicted image
+channel_titles = [
+ "Membrane",
+ "Target Phase",
+ "Predicted_Phase",
+]
+fig, axes = plt.subplots(1, 3, figsize=(30, 20))
+
+for i, sample in enumerate(test_data.test_dataloader()):
+ # Plot the phase image
+ mem_image = sample["source"]
+ channel_image = mem_image[0, 0, 0]
+ p_low, p_high = np.percentile(channel_image, (0.5, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[0].imshow(channel_image, cmap="gray")
+ axes[0].axis("off")
+ axes[0].set_title(channel_titles[0])
+
+ with torch.inference_mode(): # turn off gradient computation.
+ predicted_image = (
+ phase2fluor_model(phase_image.to(phase2fluor_model.device))
+ .cpu()
+ .numpy()
+ .squeeze(0)
+ )
+
+ target_image = sample["target"].cpu().numpy().squeeze(0)
+ # Plot the predicted images
+ channel_image = target_image[0, 0]
+ p_low, p_high = np.percentile(channel_image, (0.5, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[1].imshow(channel_image, cmap="gray")
+ axes[1].axis("off")
+ axes[1].set_title(channel_titles[1])
+
+ channel_image = predicted_image[1, 0]
+ p_low, p_high = np.percentile(channel_image, (0.1, 99.5))
+ channel_image = np.clip(channel_image, p_low, p_high)
+ axes[2].imshow(channel_image, cmap="gray")
+ axes[2].axis("off")
+ axes[2].set_title(f"VS {channel_titles[2]}")
+
+ plt.tight_layout()
+ plt.show()
+ break
# %% [markdown] tags=[]
"""
-## Checkpoint 2
-When your model finishes training, please summarize hyperparameters and performance of your models in the [this google doc](https://docs.google.com/document/d/1hZWSVRvt9KJEdYu7ib-vFBqAVQRYL8cWaP_vFznu7D8/edit#heading=h.n5u485pmzv2z)
+
Checkpoint 2
+
When your model finishes training, please summarize hyperparameters and performance of your models in the this google doc
"""
-# %%
tags=[]
-"""
-# Part 3: Tune the models.
---------------------------------------------------
-
-Learning goals: Understand how data, model capacity, and training parameters control the performance of the model. Your goal is to try to underfit or overfit the model.
-"""
-
# %% [markdown] tags=[]
"""
-### Task 3.1
+
Extra exercises
+
Tune the models and explore other architectures from VisCy
+
+
Learning goals:
+
+ - Understand how data, model capacity, and training parameters control the performance of the model. Your goal is to try to underfit or overfit the model.
+ - How can we scale it up from 2D to 3D training and predictions?
+
+
-- Choose a model you want to train (phase2fluor or fluor2phase).
-- Set up a configuration that you think will improve the performance of the model
-- Consider modifying the learning rate and see how it changes performance
-- Use training loop illustrated in previous cells to train phase2fluor and fluor2phase models to prototype your own training loop.
-- Add code to evaluate the model using Pearson Correlation and SSIM
+"""
-As your model is training, please document hyperparameters, snapshots of predictions on validation set, and loss curves for your models in [this google doc](https://docs.google.com/document/d/1hZWSVRvt9KJEdYu7ib-vFBqAVQRYL8cWaP_vFznu7D8/edit#heading=h.n5u485pmzv2z)
-
+#
+# ### Extra Example 1: Hyperparameter tuning
+#
+# - Choose a model you want to train (phase2fluor or fluor2phase).
+# - Set up a configuration that you think will improve the performance of the model
+# - Consider modifying the learning rate and see how it changes performance
+# - Use training loop illustrated in previous cells to train phase2fluor and fluor2phase models to prototype your own training loop.
+# - Add code to evaluate the model using Pearson Correlation and SSIM
+# As your model is training, please document hyperparameters, snapshots of predictions on validation set,
+# and loss curves for your models in
+# [this google doc](https://docs.google.com/document/d/1Mq-yV8FTG02xE46Mii2vzPJVYSRNdeOXkeU-EKu-irE/edit?usp=sharing)
+#
+
# %% tags=[]
##########################
######## TODO ########
@@ -875,28 +1243,28 @@ def min_max_scale(input):
##########################
######## Solution ########
##########################
+phase2fluor_config = dict(
+ in_channels=1,
+ out_channels=2,
+ encoder_blocks=[3, 3, 9, 3],
+ dims=[96, 192, 384, 768],
+ decoder_conv_blocks=2,
+ stem_kernel_size=(1, 2, 2),
+ in_stack_depth=1,
+ pretraining=False,
+)
-phase2fluor_wider_config = {
- "architecture": "2D",
- # double the number of filters at each stage
- "num_filters": [48, 96, 192, 384, 768],
- "in_channels": 1,
- "out_channels": 2,
- "residual": True,
- "dropout": 0.1,
- "task": "reg",
-}
-
-phase2fluor_wider_model = VSUNet(
- model_config=phase2fluor_wider_config.copy(),
- batch_size=BATCH_SIZE,
- loss_function=torch.nn.functional.l1_loss,
+phase2fluor_model_low_lr = VSUNet(
+ architecture="UNeXt2_2D",
+ model_config=phase2fluor_config.copy(),
+ loss_function=MixedLoss(
+ l1_alpha=0.5, l2_alpha=0.0, ms_dssim_alpha=0.5
+ ), # Changed the loss function to MixedLoss L1 and MS-SSIM
schedule="WarmupCosine",
- log_num_samples=5,
- example_input_yx_shape=YX_PATCH_SIZE,
+ lr=2e-5, # lower learning rate by factor of 10
+ log_batches_per_epoch=5, # Number of samples from each batch to log to tensorboard.
)
-
trainer = VSTrainer(
accelerator="gpu",
devices=[GPU_ID],
@@ -905,28 +1273,139 @@ def min_max_scale(input):
logger=TensorBoardLogger(
save_dir=log_dir,
name="phase2fluor",
- version="wider",
+ version="phase2fluor_low_lr",
log_graph=True,
),
fast_dev_run=True,
) # Set fast_dev_run to False to train the model.
-trainer.fit(phase2fluor_wider_model, datamodule=phase2fluor_data)
+trainer.fit(phase2fluor_model_low_lr, datamodule=phase2fluor_2D_data)
+# %% [markdown]
+"""
+
+
+Extra Example 2: 3D Virtual Staining
+
+Now, let's implement a 3D virtual staining model(Phase->Fluorescence)
+Note: This task might take longer to train +1 hr. Try it out in your free-time.
+
+
+"""
+
+# %% tags=["task"]
+data_path = Path() # TODO: Point to a 3D dataset (HEK, Neuromast)
+BATCH_SIZE = 4
+YX_PATCH_SIZE = (256, 256)
+
+phase2fluor_3D_config = ...
+
+phase2fluor_3D_data = HCSDataModule(...)
+
+phase2fluor_3D = VSUNet(...)
+
+trainer = VSTrainer(...)
+
+# Start the training
+trainer.fit(...)
# %% tags=["solution"]
##########################
######## Solution ########
##########################
+"""
+You can download the file and place it in the data folder.
+https://public.czbiohub.org/comp.micro/viscy/VSCyto3D/train/raw-and-reconstructed.zarr/
-phase2fluor_slow_model = VSUNet(
- model_config=phase2fluor_config.copy(),
+You can run the following shell script:
+```
+cd ~/data/hek3d/training
+# Download the Zarr dataset recursively (if the server supports it)
+wget -m -np -nH --cut-dirs=4 -R "index.html*" "https://public.czbiohub.org/comp.micro/viscy/VSCyto3D/train/raw-and-reconstructed.zarr/"
+```
+
+"""
+# TODO: Point to a 3D dataset (HEK, Neuromast)
+data_path = Path("./raw-and-reconstructed.zarr")
+BATCH_SIZE = 4
+YX_PATCH_SIZE = (384, 384)
+GPU_ID = 0
+n_epochs = 50
+
+## For 3D training - VSCyto3D
+source_channel = ["reconstructed-labelfree"]
+target_channel = ["reconstructed-nucleus", "reconstructed-membrane"]
+
+# Setup the new augmentations
+augmentations = [
+ RandWeightedCropd(
+ keys=source_channel + target_channel,
+ spatial_size=(-1, 512, 512),
+ num_samples=2,
+ w_key=target_channel[0],
+ ),
+ RandAffined(
+ keys=source_channel + target_channel,
+ rotate_range=[3.14, 0.0, 0.0],
+ scale_range=[0.0, 0.3, 0.3],
+ prob=0.8,
+ padding_mode="zeros",
+ shear_range=[0.0, 0.01, 0.01],
+ ),
+ RandAdjustContrastd(keys=source_channel, prob=0.5, gamma=(0.8, 1.2)),
+ RandScaleIntensityd(keys=source_channel, factors=0.5, prob=0.5),
+ RandGaussianNoised(keys=source_channel, prob=0.5, mean=0.0, std=0.3),
+ RandGaussianSmoothd(
+ keys=source_channel,
+ sigma_x=(0.25, 0.75),
+ sigma_y=(0.25, 0.75),
+ sigma_z=(0.0, 0.0),
+ prob=0.5,
+ ),
+]
+
+normalizations = [
+ NormalizeSampled(
+ keys=source_channel + target_channel,
+ level="fov_statistics",
+ subtrahend="mean",
+ divisor="std",
+ )
+]
+
+phase2fluor_3D_config = dict(
+ in_channels=1,
+ out_channels=2,
+ in_stack_depth=5,
+ backbone="convnextv2_tiny",
+ decoder_conv_blocks=2,
+ head_expansion_ratio=4,
+ stem_kernel_size=(5, 4, 4),
+)
+phase2fluor_3D_data = HCSDataModule(
+ data_path,
+ architecture="UNeXt2",
+ source_channel=source_channel,
+ target_channel=target_channel,
+ z_window_size=5,
+ split_ratio=0.8,
batch_size=BATCH_SIZE,
- loss_function=torch.nn.functional.l1_loss,
- # lower learning rate by 5 times
+ num_workers=8,
+ yx_patch_size=YX_PATCH_SIZE,
+ augmentations=augmentations,
+ normalizations=normalizations,
+)
+phase2fluor_3D_data.setup("fit")
+
+n_samples = len(phase2fluor_3D_data.train_dataset)
+steps_per_epoch = n_samples // BATCH_SIZE # steps per epoch.
+
+phase2fluor_3D = VSUNet(
+ architecture="UNeXt2",
+ model_config=phase2fluor_3D_config.copy(),
+ loss_function=MixedLoss(l1_alpha=0.5, l2_alpha=0.0, ms_dssim_alpha=0.5),
lr=2e-4,
schedule="WarmupCosine",
- log_num_samples=5,
- example_input_yx_shape=YX_PATCH_SIZE,
+ log_batches_per_epoch=5,
)
trainer = VSTrainer(
@@ -936,22 +1415,30 @@ def min_max_scale(input):
log_every_n_steps=steps_per_epoch,
logger=TensorBoardLogger(
save_dir=log_dir,
- name="phase2fluor",
- version="low_lr",
+ name="phase2fluor_3D",
+ version="3D_UNeXt2",
log_graph=True,
),
- fast_dev_run=True,
+ fast_dev_run=True, # TODO: Set to False to run full-training
)
-trainer.fit(phase2fluor_slow_model, datamodule=phase2fluor_data)
-
+trainer.fit(phase2fluor_3D, datamodule=phase2fluor_3D_data)
# %% [markdown] tags=[]
"""
-
-## Checkpoint 3
+
+
+🎉 The end of the notebook 🎉
+
Congratulations! You have trained several image translation models now!
-Please document hyperparameters, snapshots of predictions on validation set, and loss curves for your models and add the final perforance in [this google doc](https://docs.google.com/document/d/1hZWSVRvt9KJEdYu7ib-vFBqAVQRYL8cWaP_vFznu7D8/edit#heading=h.n5u485pmzv2z). We'll discuss our combined results as a group.
+
+Please remember to document the hyperparameters,
+snapshots of predictions on validation set,
+and loss curves for your models and add the final performance in
+
+this google doc
+.
+We'll discuss our combined results as a group.