| title | tags | authors | affiliations | date | bibliography | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ivadomed: A Medical Imaging Deep Learning Toolbox |
|
|
|
3 November 2020 |
paper.bib |
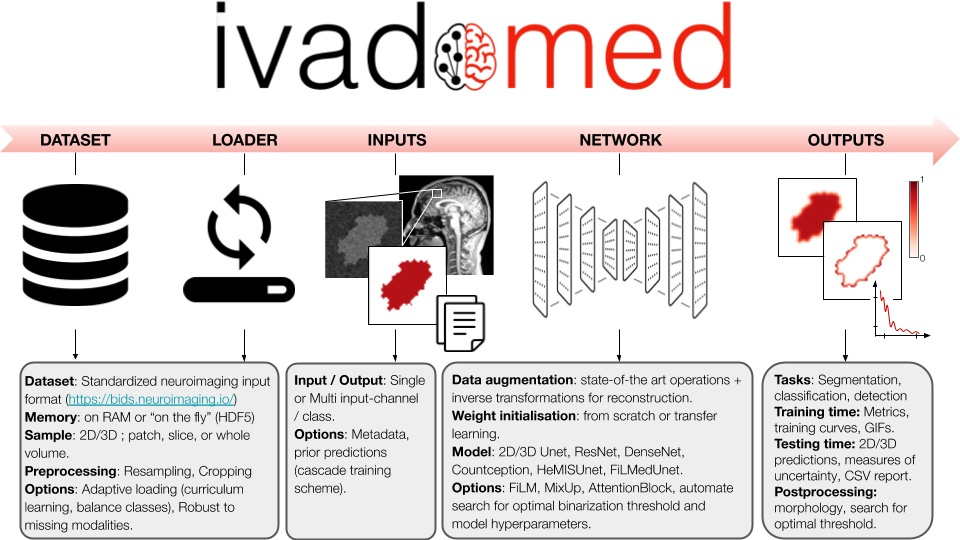
ivadomed is an open-source Python package for designing, end-to-end training, and evaluating deep learning models applied to medical imaging data. The package includes APIs, command-line tools, documentation, and tutorials. ivadomed also includes pre-trained models such as spinal tumor segmentation and vertebral labeling. Original features of ivadomed include a data loader that can parse image and subject metadata for custom data splitting or extra information during training and evaluation. Any dataset following the Brain Imaging Data Structure (BIDS) convention will be compatible with ivadomed. Beyond the traditional deep learning methods, ivadomed features cutting-edge architectures, such as FiLM [@perez2017film] and HeMis [@havaei2016hemis], as well as various uncertainty estimation methods (aleatoric and epistemic), and losses adapted to imbalanced classes and non-binary predictions. Example applications of ivadomed include MRI object detection, segmentation, and labeling of anatomical and pathological structures. ivadomed's main project page is available at https://ivadomed.org.
Deep learning applied to medical imaging presents many challenges: datasets are often not publicly-available, ground truth labels are difficult to produce, and needs are usually tailored to particular datasets and tasks [@kim_deep_2019]. There already exists a few deep learning frameworks for medical imaging, but each has their pros & cons. ivadomed notably addresses unmet needs in terms of data management, readily-available uncertainty outputs, missing modalities (in case of multi-channel training) and model comparison, to only name few of the original features.
Another challenge of medical imaging is the heterogeneity of the data across hospitals (e.g., contrast, resolution), making it difficult to create models that can generalize well. Recent cutting-edge methods address this problem, such as FiLM [@perez2017film] and HeMis [@havaei2016hemis], however they are usually not implemented within a comprehensive framework. In addition to providing these architectures, ivadomed features losses adapted to imbalanced classes and non-binary predictions.
Standard data structure: In machine learning, lots of time is spent curating the data (renaming, filtering per feature) before entering the training pipeline [@Willemink2020-au]. ivadomed features an advanced data loader compatible with a standardized data structure in neuroimaging: the Brain Imaging Data Structure (BIDS) [@bids_2016]. Thus, any dataset following the BIDS convention can readily be used by ivadomed. BIDS convention is originally designed around MRI data and accepts NIfTI file formats, but the BIDS community is actively expanding its specifications to other modalities (CT, MEG/EEG, microscopy), which ivadomed will then be able to accommodate.
Access to metadata: One benefit of the BIDS convention is that each image file is associated with a JSON file containing metadata. ivadomed's loader can parse image metadata (e.g., acquisition parameters, image contrast, resolution) and subject metadata (e.g., pathology, age, sex) for custom data splitting or extra information during training and evaluation. It is possible to modulate specific layers of a convolutional neural network using metadata information to tailor it towards a particular data domain or to enable experiments with architectures such as FiLM [@perez2017film]. Metadata could also be useful to mitigate class imbalance via data balancing techniques.
Architectures: ivadomed includes all the necessary components for training segmentation models from start to finish, including data augmentation transformations and transfer learning. Available architectures include: 2D U-Net [@Ronneberger2015unet], 3D U-Net [@isensee2017brain], ResNet [@he2016resnet], DenseNet [@Huang2017densenet], Count-ception [@Cohen2017countception], and HeMIS U-Net. These models can easily be enriched via attention blocks [@oktay2018attention] or FiLM layers (which modulate U-Net features using metadata).
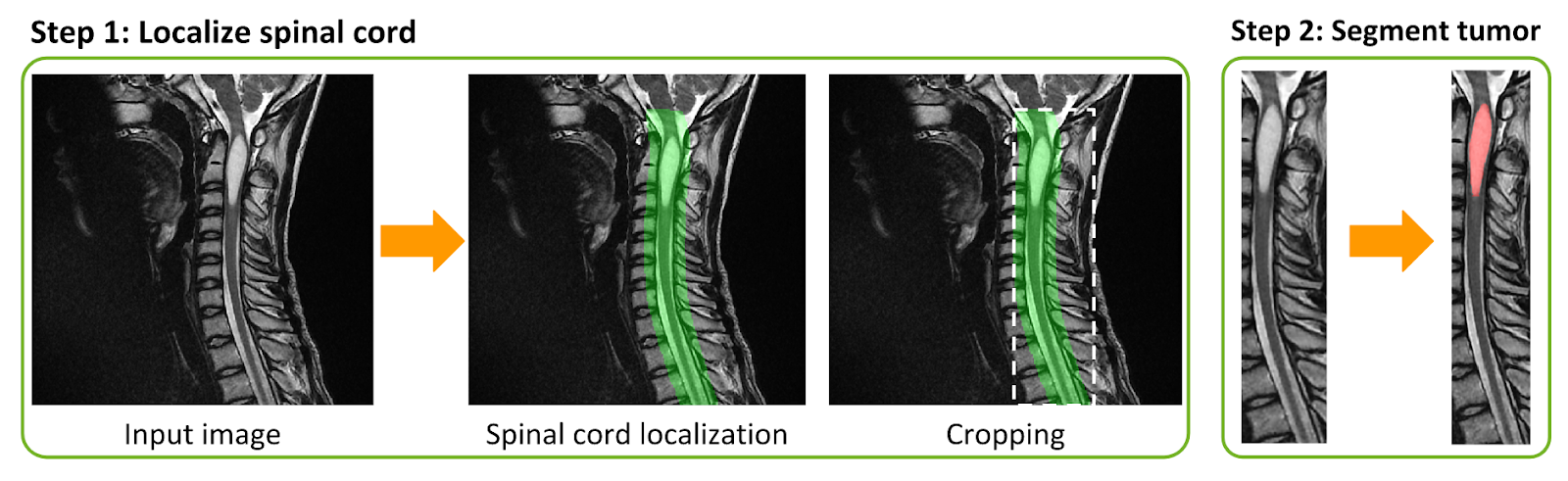
Losses and class imbalance: Popular losses are available: Dice coefficient [@milletari2016v], cross-entropy, and L2 norm, including some adapted to medical imaging challenges, such as the adaptive wing loss [@wang_adaptive_2019] for soft labels and the focal Dice loss [@wong20183d] for class imbalance. Useful to alleviate overfitting, mixup [@zhang2017mixup] was modified to handle segmentation tasks. To mitigate class imbalance, ivadomed supports cascaded architectures. With a single inference, it is possible to narrow down the region of interest via object detection and then segment a specific structure, as illustrated in \autoref{fig:lemay2020}.
Getting started: It can be overwhelming to get started and choose across all the available models, losses, and parameters. ivadomed's repository includes the script ivadomed_automate_training to configure and launch multiple trainings across GPUs. In case of interruption during training, all parameters are saved after each epoch so that training can be resumed at any time.
Uncertainty: Aleatoric [@wang_aleatoric_2019] and/or epistemic [@nair_exploring_2018] uncertainties can be computed voxel-wise and/or object-wise [@roy_quicknat_2018]. Multiple metrics are available, including entropy and coefficient of variation.
Post-processing: Predictions can be conveniently filtered using popular methods, e.g., fill holes, remove small objects, threshold using uncertainty. It is also possible to compute metrics for specific object sizes (e.g., small vs. large lesions). ivadomed has a module to find the optimal threshold value on the soft output prediction, via a grid-search finding applied to evaluation metrics or ROC curve.
Visualize performance: Convenient visualization tools are available for model design and optimization: GIF animations across training epochs, visual quality control of data augmentation, training curve plots, integration of the TensorBoard module, and output images with true/false positive labels. See this example tutorial.
Past and ongoing research projects using ivadomed are listed here. The figure below illustrates a cascaded architecture for segmenting spinal tumors on MRI data [@lemay_fully_2020].
The authors thank Ainsleigh Hill, Giselle Martel, Alexandru Jora, Nick Guenther, Joshua Newton, Christian Perone, Konstantinos Nasiotis, Valentine Louis-Lucas, Benoît Sauty-De-Chalon, Alexandru Foias and Leander Van Eekelen for their useful contributions, and Guillaume Dumas for proof-reading the manuscript. Funded by IVADO, the Canada Research Chair in Quantitative Magnetic Resonance Imaging [950-230815, 950-233166], CIHR [FDN-143263], CFI [32454, 34824], FRQS [28826], NSERC [RGPIN-2019-07244], FRQNT [2020‐RS4‐265502 UNIQUE] and TransMedTech. C.G. has a fellowship from IVADO [EX-2018-4], A.L. has a fellowship from NSERC, FRQNT and UNIQUE, O.V. has a fellowship from NSERC, FRQNT and UNIQUE, M.H.B. has a fellowship from IVADO and FRQNT.