diff --git a/.codecov.yml b/.codecov.yml
index 007de58d..70f8cec9 100644
--- a/.codecov.yml
+++ b/.codecov.yml
@@ -9,7 +9,7 @@ comment: false
flags:
numba:
paths:
- - src/cellfinder_core/detect/filters/plane/tile_walker.py
- - src/cellfinder_core/detect/filters/volume/ball_filter.py
- - src/cellfinder_core/detect/filters/volume/structure_detection.py
+ - cellfinder/core/detect/filters/plane/tile_walker.py
+ - cellfinder/core/detect/filters/volume/ball_filter.py
+ - cellfinder/core/detect/filters/volume/structure_detection.py
carryforward: true
diff --git a/.github/workflows/plugin_preview.yml b/.github/workflows/plugin_preview.yml
new file mode 100644
index 00000000..bfa67800
--- /dev/null
+++ b/.github/workflows/plugin_preview.yml
@@ -0,0 +1,19 @@
+name: napari hub Preview Page # we use this name to find your preview page artifact, so don't change it!
+# For more info on this action, see https://github.com/chanzuckerberg/napari-hub-preview-action/blob/main/action.yml
+
+on:
+ pull_request:
+
+jobs:
+ preview-page:
+ name: Preview Page Deploy
+ runs-on: ubuntu-latest
+
+ steps:
+ - name: Checkout repo
+ uses: actions/checkout@v3
+
+ - name: napari hub Preview Page Builder
+ uses: chanzuckerberg/napari-hub-preview-action@v0.1
+ with:
+ hub-ref: main
diff --git a/.github/workflows/test_and_deploy.yml b/.github/workflows/test_and_deploy.yml
index d6497728..ccbf9950 100644
--- a/.github/workflows/test_and_deploy.yml
+++ b/.github/workflows/test_and_deploy.yml
@@ -1,12 +1,23 @@
name: tests
on:
+ # Only run on pushes to main, or when version tags are pushed
push:
- branches: [ main ]
+ branches:
+ - "main"
tags:
- - '*'
+ - "v**"
+ # Run on all pull-requests
pull_request:
- branches: [ '*' ]
+ # Allow workflow dispatch from GitHub
+ workflow_dispatch:
+
+concurrency:
+ # Cancel this workflow if it is running,
+ # and then changes are applied on top of the HEAD of the branch,
+ # triggering another run of the workflow
+ group: ${{ github.workflow }}-${{ github.ref }}
+ cancel-in-progress: true
jobs:
linting:
@@ -22,6 +33,7 @@ jobs:
test:
needs: [linting, manifest]
+ name: Run package tests
runs-on: ${{ matrix.os }}
strategy:
matrix:
@@ -31,20 +43,25 @@ jobs:
# Include one windows, one macos run
include:
- os: macos-latest
- python-version: "3.9"
+ python-version: "3.10"
- os: windows-latest
- python-version: "3.9"
+ python-version: "3.10"
steps:
+ # Cache the tensorflow model so we don't have to remake it every time
- name: Cache tensorflow model
uses: actions/cache@v3
with:
path: "~/.cellfinder"
key: models-${{ hashFiles('~/.cellfinder/**') }}
+ # Setup pyqt libraries
+ - name: Setup qtpy libraries
+ uses: tlambert03/setup-qt-libs@v1
+ # Run all tests
- uses: neuroinformatics-unit/actions/test@v2
with:
python-version: ${{ matrix.python-version }}
-
+ use-xvfb: true
# Run cellfinder tests to make sure cellfinder is still compatible
# with cellfinder-core
@@ -77,7 +94,6 @@ jobs:
# Install checked out copy of cellfinder
python -m pip install .[dev]
-
- name: Run cellfinder tests
run: |
python -m pytest --color=yes -v
diff --git a/.github/workflows/test_include_guard.yaml b/.github/workflows/test_include_guard.yaml
index 5853204c..26277d98 100644
--- a/.github/workflows/test_include_guard.yaml
+++ b/.github/workflows/test_include_guard.yaml
@@ -5,7 +5,8 @@ name: Test Tensorflow include guards
on:
pull_request:
push:
- branches: [main]
+ branches:
+ - main
jobs:
tensorflow_guards:
@@ -31,7 +32,8 @@ jobs:
with:
fail-on-error: true
script: |
- import cellfinder_core
+ import cellfinder.core
+ import cellfinder.napari
- name: Uninstall tensorflow
run: python -m pip uninstall -y tensorflow
@@ -42,7 +44,7 @@ jobs:
with:
fail-on-error: false
script: |
- import cellfinder_core
+ import cellfinder.core
# exit 1 will force an actions exit with a failure reported
- name: Flag error thrown by broken import
diff --git a/.gitignore b/.gitignore
index a7202bec..7a8b1bc9 100644
--- a/.gitignore
+++ b/.gitignore
@@ -2,7 +2,7 @@
*.conf.custom
# Byte-compiled / optimized / DLL files
-__pycache__/
+**/__pycache__/
*.py[cod]
*$py.class
@@ -15,6 +15,7 @@ __pycache__/
# Distribution / packaging
.Python
+env/
build/
develop-eggs/
dist/
@@ -53,6 +54,7 @@ coverage.xml
*.cover
.hypothesis/
.pytest_cache/
+.napari_cache/
# Translations
*.mo
@@ -77,10 +79,13 @@ doc/build/
_build/
mkdocs.yml
+# MkDocs documentation
+/site/
+
# PyBuilder
target/
-# Jupyter Notebook
+# IPython Notebook
.ipynb_checkpoints
# pyenv
@@ -130,3 +135,11 @@ mprofile*.dat
benchmarks/results
benchmarks/html
benchmarks/env
+
+# OS
+.DS_Store
+
+# written by setuptools_scm
+*/_version.py
+
+.idea/
diff --git a/.napari/config.yml b/.napari/config.yml
new file mode 100644
index 00000000..adbcb0b2
--- /dev/null
+++ b/.napari/config.yml
@@ -0,0 +1,14 @@
+# Add labels from the EDAM Bioimaging ontology
+labels:
+ ontology: EDAM-BIOIMAGING:alpha06
+ terms:
+ - Image feature detection
+ - 3D image
+ - Image registration
+ - Multi-photon microscopy
+ - Light-sheet microscopy
+ - Image segmentation
+ - Image thresholding
+ - Image annotation
+ - Object classification
+ - Object feature extraction
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
index dc920786..783a3d64 100644
--- a/.pre-commit-config.yaml
+++ b/.pre-commit-config.yaml
@@ -23,15 +23,3 @@ repos:
rev: 23.7.0
hooks:
- id: black
- - repo: https://github.com/pre-commit/mirrors-mypy
- rev: v1.5.1
- hooks:
- - id: mypy
- args: [--config-file, pyproject.toml]
- exclude: benchmarks/benchmarks/tools/IO.py
- additional_dependencies:
- - types-setuptools
- - types-requests
- - types-tqdm
- - dask
- - numpy
diff --git a/CF-CORE-README.md b/CF-CORE-README.md
new file mode 100644
index 00000000..eb312390
--- /dev/null
+++ b/CF-CORE-README.md
@@ -0,0 +1,313 @@
+[](https://pypi.org/project/cellfinder-core)
+[](https://pypi.org/project/cellfinder-core)
+[](https://pepy.tech/project/cellfinder-core)
+[](https://pypi.org/project/cellfinder-core)
+[](https://github.com/brainglobe/cellfinder-core)
+[](https://github.com/brainglobe/cellfinder-core/actions)
+[](https://codecov.io/gh/brainglobe/cellfinder-core)
+[](https://github.com/python/black)
+[](https://pycqa.github.io/isort/)
+[](https://github.com/pre-commit/pre-commit)
+[](https://brainglobe.info/developers/index.html)
+[](https://twitter.com/brain_globe)
+
+# cellfinder-core
+Standalone cellfinder cell detection algorithm
+
+This package implements the cell detection algorithm from
+[Tyson, Rousseau & Niedworok et al. (2021)](https://doi.org/10.1371/journal.pcbi.1009074)
+without any dependency on data type (i.e. it can be used outside of
+whole-brain microscopy).
+
+`cellfinder-core` supports the
+[cellfinder](https://github.com/brainglobe/cellfinder) software for
+whole-brain microscopy analysis, and the algorithm can also be implemented in
+[napari](https://napari.org/index.html) using the
+[cellfinder napari plugin](https://github.com/brainglobe/cellfinder-napari).
+
+---
+
+## Instructions
+### Installation
+`cellfinder-core` supports Python >=3.9,
+and works across Linux, Windows, and should work on most versions of macOS
+(although this is not tested).
+
+Assuming you have a Python environment set up
+(e.g. [using conda](https://brainglobe.info/documentation/general/conda.html)),
+you can install `cellfinder-core` with:
+```bash
+pip install cellfinder-core
+```
+
+Once you have [installed napari](https://napari.org/index.html#installation).
+You can install napari either through the napari plugin installation tool, or
+directly from PyPI with:
+```bash
+pip install cellfinder-napari
+```
+
+N.B. To speed up cellfinder, you need CUDA & cuDNN installed. Instructions
+[here](https://brainglobe.info/documentation/general/gpu.html).
+
+#### Conda Install
+Linux and MacOS users can also install `cellfinder-core` from `conda-forge`, by running
+```sh
+conda install -c conda-forge cellfinder-core
+```
+
+Windows users can also use the command above to install `cellfinder-core`, however `tensorflow` (one of `cellfinder-core`'s core dependencies) is not available so will not be included.
+Consequentially, `cellfinder-core` will be usable - you will get `PackageNotFound` errors when attempting to import.
+To rectify this, Windows users _must_ [manually install `tensorflow`](#manual-tensorflow-installations) (and ensure their Python interpreter can see this install) for `cellfinder-core` to work.
+Please refer to the [`tensorflow` install page](https://www.tensorflow.org/install) for further guidance.
+Whether `tensorflow` is installed before or after `conda install`ing `cellfinder-core` shouldn't matter, so long as `tensorflow` is visible to the Python interpreter.
+
+### Usage
+Before using cellfinder-core, it may be useful to take a look at the
+[paper](https://doi.org/10.1371/journal.pcbi.1009074) which
+outlines the algorithm.
+
+The API is not yet fully documented. For an idea of what the parameters do,
+see the documentation for the cellfinder whole-brain microscopy image analysis
+command-line tool ([cell candidate detection](https://brainglobe.info/documentation/cellfinder/user-guide/command-line/candidate-detection.html),
+[cell candidate classification](https://brainglobe.info/documentation/cellfinder/user-guide/command-line/classification.html)).
+It may also be useful to try the
+[cellfinder napari plugin](https://github.com/brainglobe/cellfinder-napari)
+so you can adjust the parameters in a GUI.
+
+#### To run the full pipeline (cell candidate detection and classification)
+```python
+from cellfinder.core.main import main as cellfinder_run
+import tifffile
+
+signal_array = tifffile.imread("/path/to/signal_image.tif")
+background_array = tifffile.imread("/path/to/background_image.tif")
+
+voxel_sizes = [5, 2, 2] # in microns
+detected_cells = cellfinder_run(signal_array,background_array,voxel_sizes)
+```
+
+The output is a list of
+[brainglobe-utils Cell objects](https://github.com/brainglobe/brainglobe-utils/blob/044a735049c1323466d277f9df1c3abad8b2bb8d/brainglobe_utils/cells/cells.py#L19)
+Each `Cell` has a centroid coordinate, and a type:
+
+```python
+print(detected_cells[0])
+# Cell: x: 132, y: 308, z: 10, type: 2
+```
+
+Cell type 2 is a "real" cell, and Cell type 1 is a "rejected" object (i.e.
+not classified as a cell):
+
+```python
+from brainglobe_utils.cells.cells import Cell
+print(Cell.CELL)
+# 2
+
+print(Cell.NO_CELL)
+# 1
+```
+
+#### Saving the results
+If you want to save the detected cells for use in other BrainGlobe software (e.g. the
+[cellfinder napari plugin](https://brainglobe.info/documentation/cellfinder/user-guide/napari-plugin/index.html)),
+you can save in the cellfinder XML standard:
+```python
+from brainglobe_utils.IO.cells import save_cells
+save_cells(detected_cells, "/path/to/cells.xml")
+```
+You can load these back with:
+```python
+from brainglobe_utils.IO.cells import get_cells
+cells = get_cells("/path/to/cells.xml")
+```
+
+
+#### Using dask for lazy loading
+`cellfinder-core` supports most array-like objects. Using
+[Dask arrays](https://docs.dask.org/en/latest/array.html) allows for lazy
+loading of data, allowing large (e.g. TB) datasets to be processed.
+`cellfinder-core` comes with a function
+(based on [napari-ndtiffs](https://github.com/tlambert03/napari-ndtiffs)) to
+load a series of image files (e.g. a directory of 2D tiff files) as a Dask
+array. `cellfinder-core` can then be used in the same way as with a numpy array.
+
+```python
+from cellfinder.core.main import main as cellfinder_run
+from cellfinder.core.tools.IO import read_with_dask
+
+signal_array = read_with_dask("/path/to/signal_image_directory")
+background_array = read_with_dask("/path/to/background_image_directory")
+
+voxel_sizes = [5, 2, 2] # in microns
+detected_cells = cellfinder_run(signal_array,background_array,voxel_sizes)
+
+```
+
+#### Running the cell candidate detection and classification separately.
+```python
+import tifffile
+from pathlib import Path
+
+from cellfinder.core.detect import detect
+from cellfinder.core.classify import classify
+from cellfinder.core.tools.prep import prep_classification
+
+signal_array = tifffile.imread("/path/to/signal_image.tif")
+background_array = tifffile.imread("/path/to/background_image.tif")
+voxel_sizes = [5, 2, 2] # in microns
+
+home = Path.home()
+install_path = home / ".cellfinder" # default
+
+start_plane=0
+end_plane=-1
+trained_model=None
+model_weights=None
+model="resnet50_tv"
+batch_size=32
+n_free_cpus=2
+network_voxel_sizes=[5, 1, 1]
+soma_diameter=16
+ball_xy_size=6
+ball_z_size=15
+ball_overlap_fraction=0.6
+log_sigma_size=0.2
+n_sds_above_mean_thresh=10
+soma_spread_factor=1.4
+max_cluster_size=100000
+cube_width=50
+cube_height=50
+cube_depth=20
+network_depth="50"
+
+model_weights = prep_classification(
+ trained_model, model_weights, install_path, model, n_free_cpus
+)
+
+cell_candidates = detect.main(
+ signal_array,
+ start_plane,
+ end_plane,
+ voxel_sizes,
+ soma_diameter,
+ max_cluster_size,
+ ball_xy_size,
+ ball_z_size,
+ ball_overlap_fraction,
+ soma_spread_factor,
+ n_free_cpus,
+ log_sigma_size,
+ n_sds_above_mean_thresh,
+)
+
+if len(cell_candidates) > 0: # Don't run if there's nothing to classify
+ classified_cells = classify.main(
+ cell_candidates,
+ signal_array,
+ background_array,
+ n_free_cpus,
+ voxel_sizes,
+ network_voxel_sizes,
+ batch_size,
+ cube_height,
+ cube_width,
+ cube_depth,
+ trained_model,
+ model_weights,
+ network_depth,
+ )
+```
+#### Training the network
+The training data needed are matched pairs (signal & background) of small
+(usually 50 x 50 x 100um) images centered on the coordinate of candidate cells.
+These can be generated however you like, but I recommend using the
+[Napari plugin](https://brainglobe.
+info/documentation/cellfinder/user-guide/napari-plugin/training-data-generation.html).
+
+`cellfinder-core` comes with a 50-layer ResNet trained on ~100,000 data points
+from serial two-photon microscopy images of mouse brains
+(available [here](https://gin.g-node.org/cellfinder/training_data)).
+
+Training the network is likely simpler using the
+[command-line interface](https://brainglobe.info/documentation/cellfinder/user-guide/command-line/training/index.html)
+or the [Napari plugin](https://brainglobe.info/documentation/cellfinder/user-guide/napari-plugin/training-the-network.html),
+but it is possible through the Python API.
+
+```python
+from pathlib import Path
+from cellfinder.core.train.train_yml import run as run_training
+
+# list of training yml files
+yaml_files = [Path("/path/to/training_yml.yml)]
+
+# where to save the output
+output_directory = Path("/path/to/saved_training_data")
+
+home = Path.home()
+install_path = home / ".cellfinder" # default
+
+run_training(
+ output_directory,
+ yaml_files,
+ install_path=install_path,
+ learning_rate=0.0001,
+ continue_training=True, # by default use supplied model
+ test_fraction=0.1,
+ batch_size=32,
+ save_progress=True,
+ epochs=10,
+)
+```
+
+---
+### More info
+
+More documentation about cellfinder and other BrainGlobe tools can be
+found [here](https://brainglobe.info).
+
+This software is at a very early stage, and was written with our data in mind.
+Over time we hope to support other data types/formats. If you have any
+questions or issues, please get in touch [on the forum](https://forum.image.sc/tag/brainglobe) or by
+[raising an issue](https://github.com/brainglobe/cellfinder-core/issues).
+
+---
+## Illustration
+
+### Introduction
+cellfinder takes a stitched, but otherwise raw dataset with at least
+two channels:
+ * Background channel (i.e. autofluorescence)
+ * Signal channel, the one with the cells to be detected:
+
+
+**Raw coronal serial two-photon mouse brain image showing labelled cells**
+
+
+### Cell candidate detection
+Classical image analysis (e.g. filters, thresholding) is used to find
+cell-like objects (with false positives):
+
+
+**Candidate cells (including many artefacts)**
+
+
+### Cell candidate classification
+A deep-learning network (ResNet) is used to classify cell candidates as true
+cells or artefacts:
+
+
+**Cassified cell candidates. Yellow - cells, Blue - artefacts**
+
+## Contributing
+Contributions to cellfinder-core are more than welcome. Please see the [developers guide](https://brainglobe.info/developers/index.html).
+
+---
+## Citing cellfinder
+If you find this plugin useful, and use it in your research, please cite the paper outlining the cell detection algorithm:
+> Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074
+[https://doi.org/10.1371/journal.pcbi.1009074](https://doi.org/10.1371/journal.pcbi.1009074)
+
+**If you use this, or any other tools in the brainglobe suite, please
+ [let us know](mailto:hello@brainglobe.info?subject=cellfinder-core), and
+ we'd be happy to promote your paper/talk etc.**
diff --git a/CF-NAPARI-README.md b/CF-NAPARI-README.md
new file mode 100644
index 00000000..fcd853ba
--- /dev/null
+++ b/CF-NAPARI-README.md
@@ -0,0 +1,92 @@
+# cellfinder-napari
+
+[](https://github.com/napari/cellfinder-napari/raw/master/LICENSE)
+[](https://pypi.org/project/cellfinder-napari)
+[](https://python.org)
+[](https://github.com/brainglobe/cellfinder-napari/actions)
+[](https://codecov.io/gh/brainglobe/cellfinder-napari)
+[](https://pepy.tech/project/cellfinder-napari)
+[](https://pypi.org/project/cellfinder)
+[](https://github.com/brainglobe/cellfinder-napari)
+[](https://github.com/python/black)
+[](https://pycqa.github.io/isort/)
+[](https://github.com/pre-commit/pre-commit)
+[](https://docs.brainglobe.info/cellfinder/contributing)
+[](https://brainglobe.info/documentation/cellfinder/index.html)
+[](https://twitter.com/brain_globe)
+
+### Efficient cell detection in large images (e.g. whole mouse brain images)
+
+`cellfinder-napari` is a front-end to [cellfinder-core](https://github.com/brainglobe/cellfinder-core) to allow ease of use within the [napari](https://napari.org/index.html) multidimensional image viewer. For more details on this approach, please see [Tyson, Rousseau & Niedworok et al. (2021)](https://doi.org/10.1371/journal.pcbi.1009074). This algorithm can also be used within the original
+[cellfinder](https://github.com/brainglobe/cellfinder) software for
+whole-brain microscopy analysis.
+
+`cellfinder-napari`, `cellfinder` and `cellfinder-core` were developed by [Charly Rousseau](https://github.com/crousseau) and [Adam Tyson](https://github.com/adamltyson) in the [Margrie Lab](https://www.sainsburywellcome.org/web/groups/margrie-lab), based on previous work by [Christian Niedworok](https://github.com/cniedwor), generously supported by the [Sainsbury Wellcome Centre](https://www.sainsburywellcome.org/web/).
+
+----
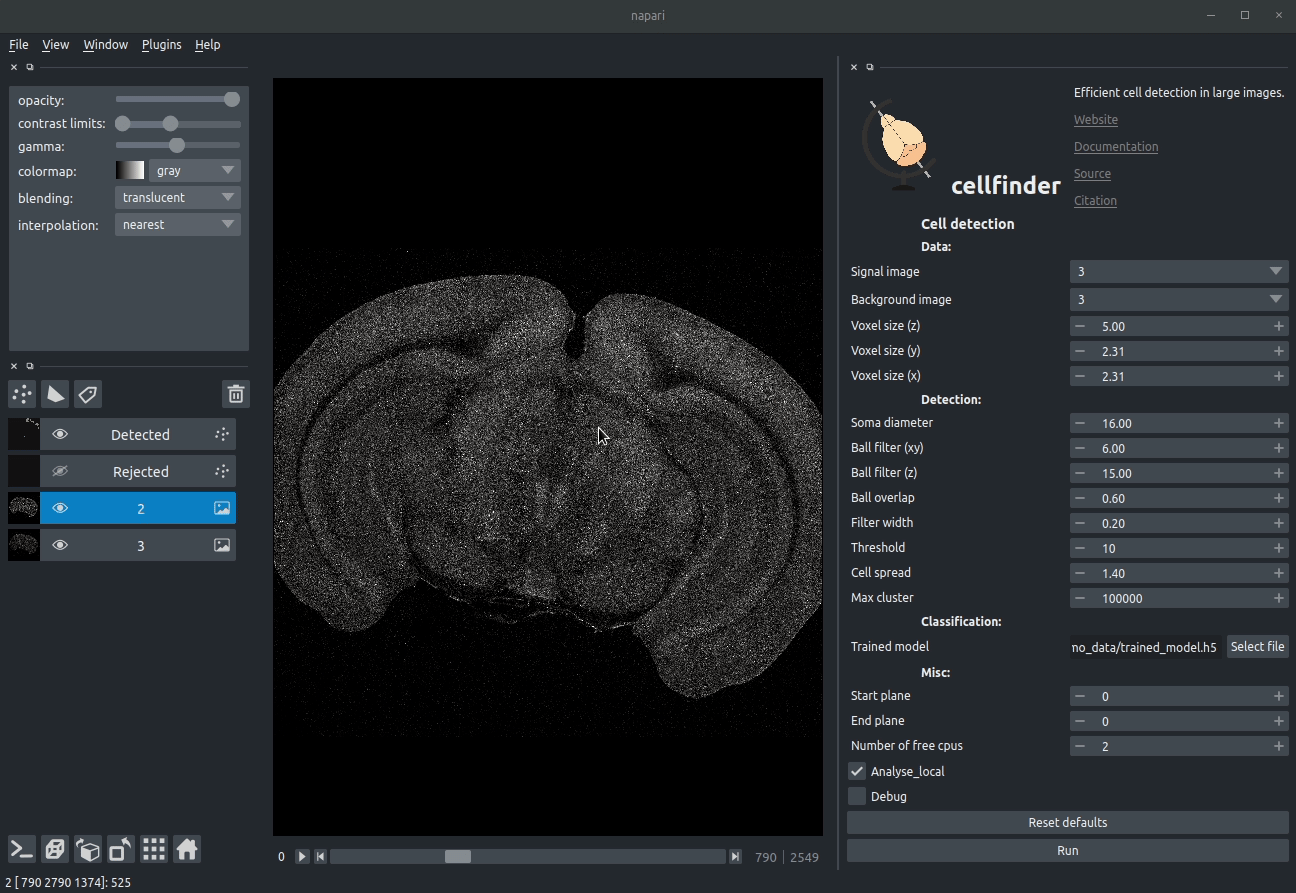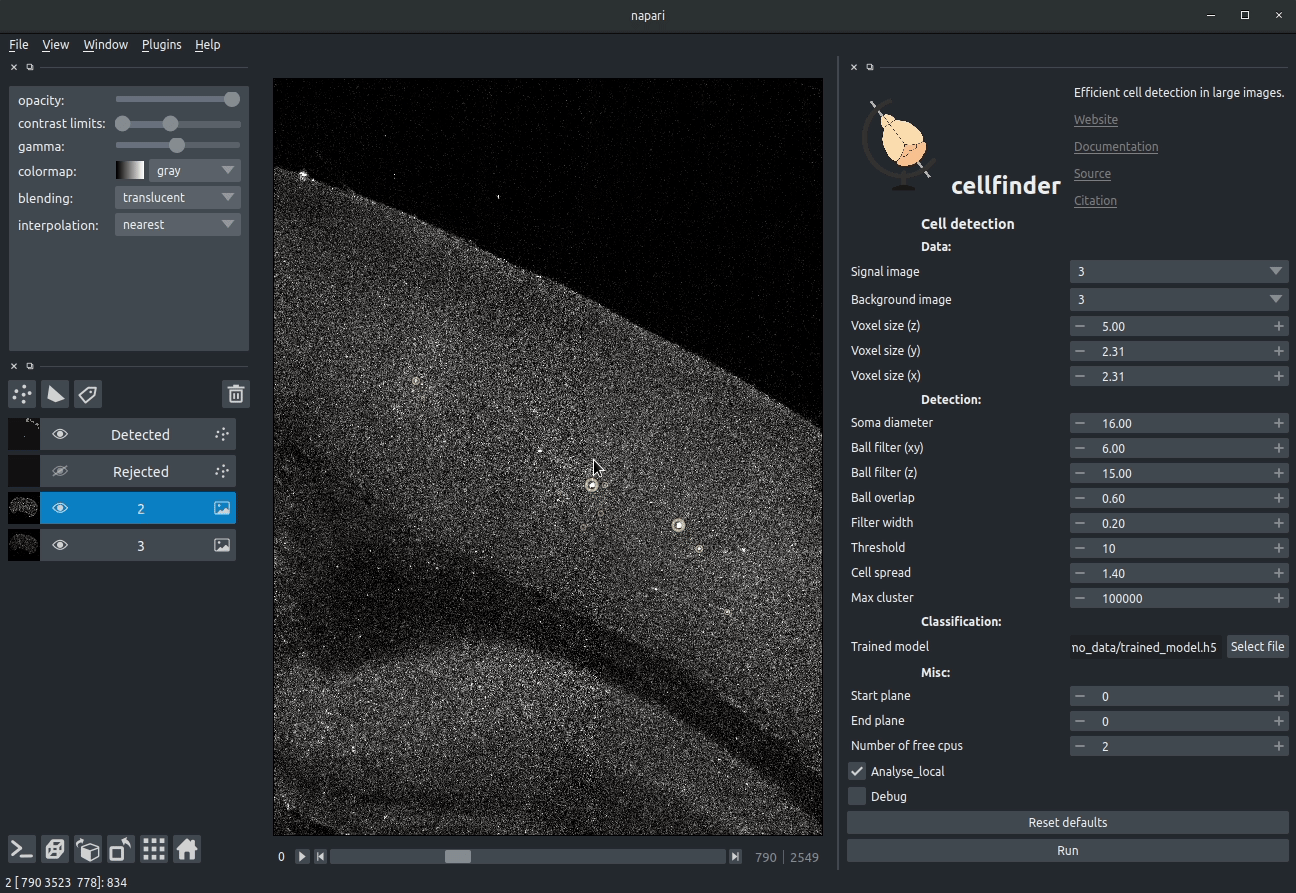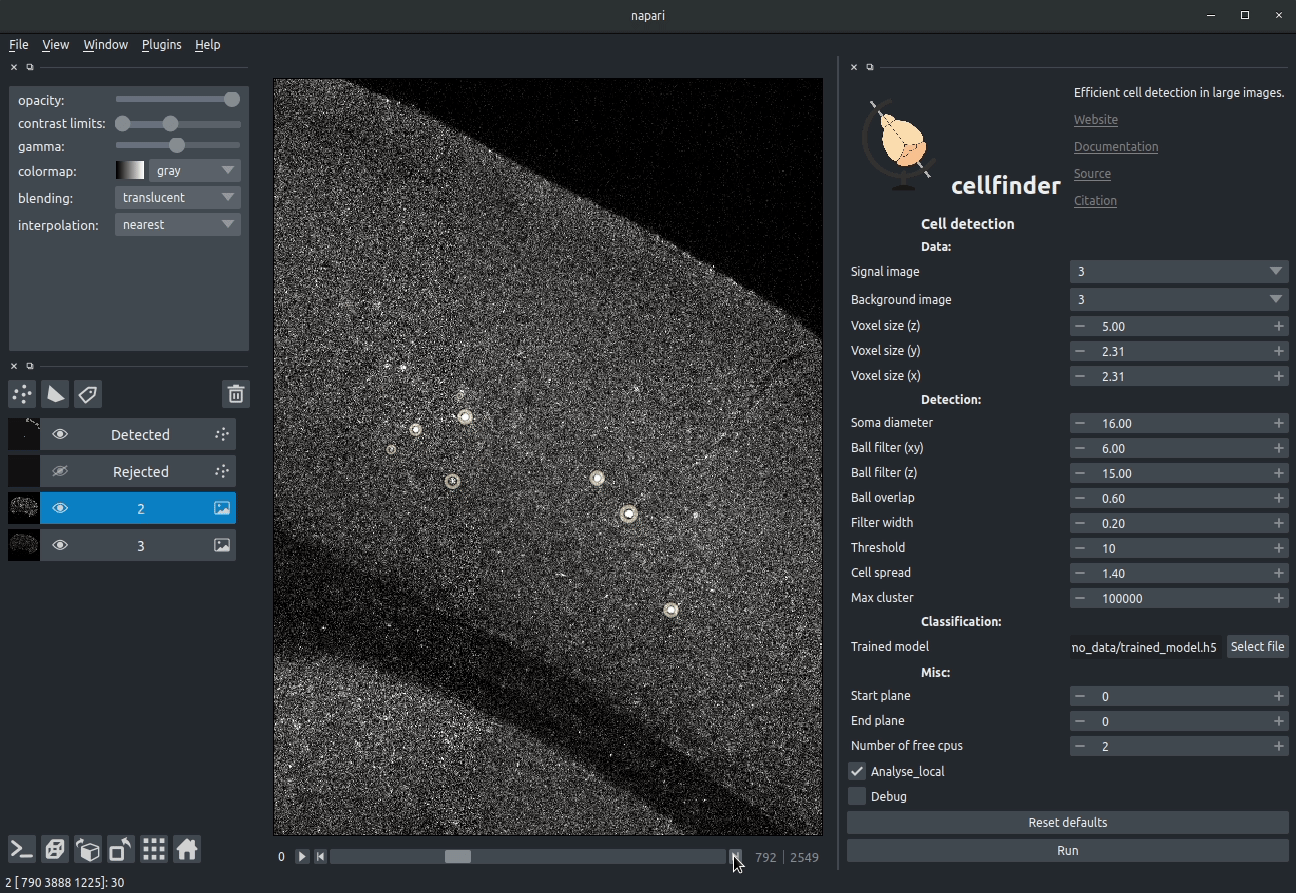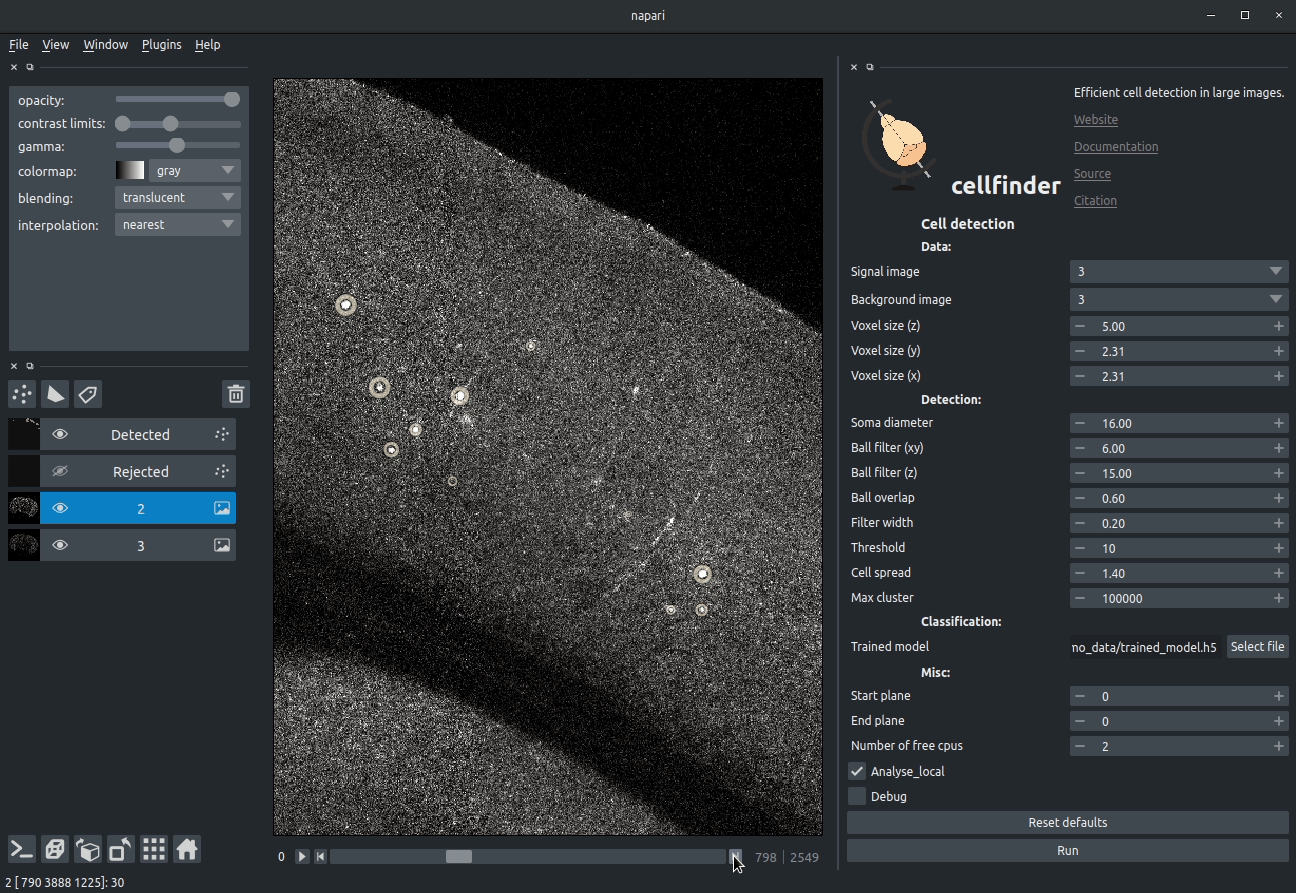
+
+
+**Visualising detected cells in the cellfinder napari plugin**
+
+----
+## Instructions
+
+### Installation
+Once you have [installed napari](https://napari.org/index.html#installation).
+You can install napari either through the napari plugin installation tool, or
+directly from PyPI with:
+```bash
+pip install cellfinder-napari
+```
+
+### Usage
+Full documentation can be
+found [here](https://brainglobe.info/documentation/cellfinder/index.html).
+
+This software is at a very early stage, and was written with our data in mind.
+Over time we hope to support other data types/formats. If you have any
+questions or issues, please get in touch [on the forum](https://forum.image.sc/tag/brainglobe) or by
+[raising an issue](https://github.com/brainglobe/cellfinder-napari/issues).
+
+
+---
+## Illustration
+
+### Introduction
+cellfinder takes a stitched, but otherwise raw dataset with at least
+two channels:
+ * Background channel (i.e. autofluorescence)
+ * Signal channel, the one with the cells to be detected:
+
+
+**Raw coronal serial two-photon mouse brain image showing labelled cells**
+
+
+### Cell candidate detection
+Classical image analysis (e.g. filters, thresholding) is used to find
+cell-like objects (with false positives):
+
+
+**Candidate cells (including many artefacts)**
+
+
+### Cell candidate classification
+A deep-learning network (ResNet) is used to classify cell candidates as true
+cells or artefacts:
+
+
+**Cassified cell candidates. Yellow - cells, Blue - artefacts**
+
+## Contributing
+Contributions to cellfinder-napari are more than welcome. Please see the [developers guide](https://brainglobe.info/developers/index.html).
+
+## Citing cellfinder
+
+If you find this plugin useful, and use it in your research, please cite the paper outlining the cell detection algorithm:
+> Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074
+[https://doi.org/10.1371/journal.pcbi.1009074](https://doi.org/10.1371/journal.pcbi.1009074)
+
+
+**If you use this, or any other tools in the brainglobe suite, please
+ [let us know](mailto:code@adamltyson.com?subject=cellfinder-napari), and
+ we'd be happy to promote your paper/talk etc.**
diff --git a/CHANGELOG.md b/CORE_CHANGELOG.md
similarity index 100%
rename from CHANGELOG.md
rename to CORE_CHANGELOG.md
diff --git a/LICENSE b/LICENSE
index 3e221c2a..18ba1208 100644
--- a/LICENSE
+++ b/LICENSE
@@ -1,6 +1,6 @@
BSD 3-Clause License
-Copyright (c) 2020, University College London
+Copyright (c) 2020, Adam Tyson, University College London
All rights reserved.
Redistribution and use in source and binary forms, with or without
@@ -13,8 +13,8 @@ modification, are permitted provided that the following conditions are met:
this list of conditions and the following disclaimer in the documentation
and/or other materials provided with the distribution.
-* Neither the name of the copyright holder nor the names of its
- contributors may be used to endorse or promote products derived from
+* Neither the name of the copyright holder, nor the names of its
+ contributors, nor the name cellfinder, may be used to endorse or promote products derived from
this software without specific prior written permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
diff --git a/MANIFEST.in b/MANIFEST.in
index 995fdf64..c4e738ec 100644
--- a/MANIFEST.in
+++ b/MANIFEST.in
@@ -1,13 +1,19 @@
-include README.md
+include .napari/config.yml
include LICENSE
-include pyproject.toml
+include README.md
+include requirements.txt
+include CF-CORE-README.md
+include CF-NAPARI-README.md
-exclude *.yml
-exclude *.yaml
+exclude .pre-commit-config.yaml
+exclude .codecov.yml
+exclude CORE_CHANGELOG.md
exclude tox.ini
-exclude CHANGELOG.md
-graft src
+graft cellfinder
+include cellfinder/napari/napari.yaml
prune benchmarks
+prune examples
+prune resources
prune tests
diff --git a/README.md b/README.md
index cf505ceb..4c9a0ac6 100644
--- a/README.md
+++ b/README.md
@@ -1,313 +1,3 @@
-[](https://pypi.org/project/cellfinder-core)
-[](https://pypi.org/project/cellfinder-core)
-[](https://pepy.tech/project/cellfinder-core)
-[](https://pypi.org/project/cellfinder-core)
-[](https://github.com/brainglobe/cellfinder-core)
-[](https://github.com/brainglobe/cellfinder-core/actions)
-[](https://codecov.io/gh/brainglobe/cellfinder-core)
-[](https://github.com/python/black)
-[](https://pycqa.github.io/isort/)
-[](https://github.com/pre-commit/pre-commit)
-[](https://brainglobe.info/developers/index.html)
-[](https://twitter.com/brain_globe)
+# `cellfinder`
-# cellfinder-core
-Standalone cellfinder cell detection algorithm
-
-This package implements the cell detection algorithm from
-[Tyson, Rousseau & Niedworok et al. (2021)](https://doi.org/10.1371/journal.pcbi.1009074)
-without any dependency on data type (i.e. it can be used outside of
-whole-brain microscopy).
-
-`cellfinder-core` supports the
-[cellfinder](https://github.com/brainglobe/cellfinder) software for
-whole-brain microscopy analysis, and the algorithm can also be implemented in
-[napari](https://napari.org/index.html) using the
-[cellfinder napari plugin](https://github.com/brainglobe/cellfinder-napari).
-
----
-
-## Instructions
-### Installation
-`cellfinder-core` supports Python >=3.9,
-and works across Linux, Windows, and should work on most versions of macOS
-(although this is not tested).
-
-Assuming you have a Python environment set up
-(e.g. [using conda](https://brainglobe.info/documentation/general/conda.html)),
-you can install `cellfinder-core` with:
-```bash
-pip install cellfinder-core
-```
-
-Once you have [installed napari](https://napari.org/index.html#installation).
-You can install napari either through the napari plugin installation tool, or
-directly from PyPI with:
-```bash
-pip install cellfinder-napari
-```
-
-N.B. To speed up cellfinder, you need CUDA & cuDNN installed. Instructions
-[here](https://brainglobe.info/documentation/general/gpu.html).
-
-#### Conda Install
-Linux and MacOS users can also install `cellfinder-core` from `conda-forge`, by running
-```sh
-conda install -c conda-forge cellfinder-core
-```
-
-Windows users can also use the command above to install `cellfinder-core`, however `tensorflow` (one of `cellfinder-core`'s core dependencies) is not available so will not be included.
-Consequentially, `cellfinder-core` will be usable - you will get `PackageNotFound` errors when attempting to import.
-To rectify this, Windows users _must_ [manually install `tensorflow`](#manual-tensorflow-installations) (and ensure their Python interpreter can see this install) for `cellfinder-core` to work.
-Please refer to the [`tensorflow` install page](https://www.tensorflow.org/install) for further guidance.
-Whether `tensorflow` is installed before or after `conda install`ing `cellfinder-core` shouldn't matter, so long as `tensorflow` is visible to the Python interpreter.
-
-### Usage
-Before using cellfinder-core, it may be useful to take a look at the
-[paper](https://doi.org/10.1371/journal.pcbi.1009074) which
-outlines the algorithm.
-
-The API is not yet fully documented. For an idea of what the parameters do,
-see the documentation for the cellfinder whole-brain microscopy image analysis
-command-line tool ([cell candidate detection](https://brainglobe.info/documentation/cellfinder/user-guide/command-line/candidate-detection.html),
-[cell candidate classification](https://brainglobe.info/documentation/cellfinder/user-guide/command-line/classification.html)).
-It may also be useful to try the
-[cellfinder napari plugin](https://github.com/brainglobe/cellfinder-napari)
-so you can adjust the parameters in a GUI.
-
-#### To run the full pipeline (cell candidate detection and classification)
-```python
-from cellfinder_core.main import main as cellfinder_run
-import tifffile
-
-signal_array = tifffile.imread("/path/to/signal_image.tif")
-background_array = tifffile.imread("/path/to/background_image.tif")
-
-voxel_sizes = [5, 2, 2] # in microns
-detected_cells = cellfinder_run(signal_array,background_array,voxel_sizes)
-```
-
-The output is a list of
-[brainglobe-utils Cell objects](https://github.com/brainglobe/brainglobe-utils/blob/044a735049c1323466d277f9df1c3abad8b2bb8d/brainglobe_utils/cells/cells.py#L19)
-Each `Cell` has a centroid coordinate, and a type:
-
-```python
-print(detected_cells[0])
-# Cell: x: 132, y: 308, z: 10, type: 2
-```
-
-Cell type 2 is a "real" cell, and Cell type 1 is a "rejected" object (i.e.
-not classified as a cell):
-
-```python
-from brainglobe_utils.cells.cells import Cell
-print(Cell.CELL)
-# 2
-
-print(Cell.NO_CELL)
-# 1
-```
-
-#### Saving the results
-If you want to save the detected cells for use in other BrainGlobe software (e.g. the
-[cellfinder napari plugin](https://brainglobe.info/documentation/cellfinder/user-guide/napari-plugin/index.html)),
-you can save in the cellfinder XML standard:
-```python
-from brainglobe_utils.IO.cells import save_cells
-save_cells(detected_cells, "/path/to/cells.xml")
-```
-You can load these back with:
-```python
-from brainglobe_utils.IO.cells import get_cells
-cells = get_cells("/path/to/cells.xml")
-```
-
-
-#### Using dask for lazy loading
-`cellfinder-core` supports most array-like objects. Using
-[Dask arrays](https://docs.dask.org/en/latest/array.html) allows for lazy
-loading of data, allowing large (e.g. TB) datasets to be processed.
-`cellfinder-core` comes with a function
-(based on [napari-ndtiffs](https://github.com/tlambert03/napari-ndtiffs)) to
-load a series of image files (e.g. a directory of 2D tiff files) as a Dask
-array. `cellfinder-core` can then be used in the same way as with a numpy array.
-
-```python
-from cellfinder_core.main import main as cellfinder_run
-from cellfinder_core.tools.IO import read_with_dask
-
-signal_array = read_with_dask("/path/to/signal_image_directory")
-background_array = read_with_dask("/path/to/background_image_directory")
-
-voxel_sizes = [5, 2, 2] # in microns
-detected_cells = cellfinder_run(signal_array,background_array,voxel_sizes)
-
-```
-
-#### Running the cell candidate detection and classification separately.
-```python
-import tifffile
-from pathlib import Path
-
-from cellfinder_core.detect import detect
-from cellfinder_core.classify import classify
-from cellfinder_core.tools.prep import prep_classification
-
-signal_array = tifffile.imread("/path/to/signal_image.tif")
-background_array = tifffile.imread("/path/to/background_image.tif")
-voxel_sizes = [5, 2, 2] # in microns
-
-home = Path.home()
-install_path = home / ".cellfinder" # default
-
-start_plane=0
-end_plane=-1
-trained_model=None
-model_weights=None
-model="resnet50_tv"
-batch_size=32
-n_free_cpus=2
-network_voxel_sizes=[5, 1, 1]
-soma_diameter=16
-ball_xy_size=6
-ball_z_size=15
-ball_overlap_fraction=0.6
-log_sigma_size=0.2
-n_sds_above_mean_thresh=10
-soma_spread_factor=1.4
-max_cluster_size=100000
-cube_width=50
-cube_height=50
-cube_depth=20
-network_depth="50"
-
-model_weights = prep_classification(
- trained_model, model_weights, install_path, model, n_free_cpus
-)
-
-cell_candidates = detect.main(
- signal_array,
- start_plane,
- end_plane,
- voxel_sizes,
- soma_diameter,
- max_cluster_size,
- ball_xy_size,
- ball_z_size,
- ball_overlap_fraction,
- soma_spread_factor,
- n_free_cpus,
- log_sigma_size,
- n_sds_above_mean_thresh,
-)
-
-if len(cell_candidates) > 0: # Don't run if there's nothing to classify
- classified_cells = classify.main(
- cell_candidates,
- signal_array,
- background_array,
- n_free_cpus,
- voxel_sizes,
- network_voxel_sizes,
- batch_size,
- cube_height,
- cube_width,
- cube_depth,
- trained_model,
- model_weights,
- network_depth,
- )
-```
-#### Training the network
-The training data needed are matched pairs (signal & background) of small
-(usually 50 x 50 x 100um) images centered on the coordinate of candidate cells.
-These can be generated however you like, but I recommend using the
-[Napari plugin](https://brainglobe.
-info/documentation/cellfinder/user-guide/napari-plugin/training-data-generation.html).
-
-`cellfinder-core` comes with a 50-layer ResNet trained on ~100,000 data points
-from serial two-photon microscopy images of mouse brains
-(available [here](https://gin.g-node.org/cellfinder/training_data)).
-
-Training the network is likely simpler using the
-[command-line interface](https://brainglobe.info/documentation/cellfinder/user-guide/command-line/training/index.html)
-or the [Napari plugin](https://brainglobe.info/documentation/cellfinder/user-guide/napari-plugin/training-the-network.html),
-but it is possible through the Python API.
-
-```python
-from pathlib import Path
-from cellfinder_core.train.train_yml import run as run_training
-
-# list of training yml files
-yaml_files = [Path("/path/to/training_yml.yml)]
-
-# where to save the output
-output_directory = Path("/path/to/saved_training_data")
-
-home = Path.home()
-install_path = home / ".cellfinder" # default
-
-run_training(
- output_directory,
- yaml_files,
- install_path=install_path,
- learning_rate=0.0001,
- continue_training=True, # by default use supplied model
- test_fraction=0.1,
- batch_size=32,
- save_progress=True,
- epochs=10,
-)
-```
-
----
-### More info
-
-More documentation about cellfinder and other BrainGlobe tools can be
-found [here](https://brainglobe.info).
-
-This software is at a very early stage, and was written with our data in mind.
-Over time we hope to support other data types/formats. If you have any
-questions or issues, please get in touch [on the forum](https://forum.image.sc/tag/brainglobe) or by
-[raising an issue](https://github.com/brainglobe/cellfinder-core/issues).
-
----
-## Illustration
-
-### Introduction
-cellfinder takes a stitched, but otherwise raw dataset with at least
-two channels:
- * Background channel (i.e. autofluorescence)
- * Signal channel, the one with the cells to be detected:
-
-
-**Raw coronal serial two-photon mouse brain image showing labelled cells**
-
-
-### Cell candidate detection
-Classical image analysis (e.g. filters, thresholding) is used to find
-cell-like objects (with false positives):
-
-
-**Candidate cells (including many artefacts)**
-
-
-### Cell candidate classification
-A deep-learning network (ResNet) is used to classify cell candidates as true
-cells or artefacts:
-
-
-**Cassified cell candidates. Yellow - cells, Blue - artefacts**
-
-## Contributing
-Contributions to cellfinder-core are more than welcome. Please see the [developers guide](https://brainglobe.info/developers/index.html).
-
----
-## Citing cellfinder
-If you find this plugin useful, and use it in your research, please cite the paper outlining the cell detection algorithm:
-> Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074
-[https://doi.org/10.1371/journal.pcbi.1009074](https://doi.org/10.1371/journal.pcbi.1009074)
-
-**If you use this, or any other tools in the brainglobe suite, please
- [let us know](mailto:hello@brainglobe.info?subject=cellfinder-core), and
- we'd be happy to promote your paper/talk etc.**
+The README needs to be updated, but the original two are preserved in the top level files.
diff --git a/benchmarks/benchmarks/imports.py b/benchmarks/benchmarks/imports.py
index d7ef25cc..e7356cdd 100644
--- a/benchmarks/benchmarks/imports.py
+++ b/benchmarks/benchmarks/imports.py
@@ -3,41 +3,41 @@
# ------------------------------------
def timeraw_import_main():
return """
- from cellfinder_core.main import main
+ from cellfinder.core.main import main
"""
def timeraw_import_io_dask():
return """
- from cellfinder_core.tools.IO import read_with_dask
+ from cellfinder.core.tools.IO import read_with_dask
"""
def timeraw_import_io_tiff_meta():
return """
- from cellfinder_core.tools.IO import get_tiff_meta
+ from cellfinder.core.tools.IO import get_tiff_meta
"""
def timeraw_import_prep_tensorflow():
return """
- from cellfinder_core.tools.prep import prep_tensorflow
+ from cellfinder.core.tools.prep import prep_tensorflow
"""
def timeraw_import_prep_models():
return """
- from cellfinder_core.tools.prep import prep_models
+ from cellfinder.core.tools.prep import prep_models
"""
def timeraw_import_prep_classification():
return """
- from cellfinder_core.tools.prep import prep_classification
+ from cellfinder.core.tools.prep import prep_classification
"""
def timeraw_import_prep_training():
return """
- from cellfinder_core.tools.prep import prep_training
+ from cellfinder.core.tools.prep import prep_training
"""
diff --git a/benchmarks/benchmarks/tools/IO.py b/benchmarks/benchmarks/tools/IO.py
index 6bc56057..57d3733f 100644
--- a/benchmarks/benchmarks/tools/IO.py
+++ b/benchmarks/benchmarks/tools/IO.py
@@ -1,6 +1,6 @@
from pathlib import Path
-from cellfinder_core.tools.IO import get_tiff_meta, read_with_dask
+from cellfinder.core.tools.IO import get_tiff_meta, read_with_dask
CELLFINDER_CORE_PATH = Path(__file__).parents[3]
TESTS_DATA_INTEGRATION_PATH = (
diff --git a/benchmarks/benchmarks/tools/prep.py b/benchmarks/benchmarks/tools/prep.py
index 09e1e755..27d9620b 100644
--- a/benchmarks/benchmarks/tools/prep.py
+++ b/benchmarks/benchmarks/tools/prep.py
@@ -3,7 +3,7 @@
from brainglobe_utils.general.system import get_num_processes
-from cellfinder_core.tools.prep import (
+from cellfinder.core.tools.prep import (
prep_classification,
prep_models,
prep_tensorflow,
diff --git a/benchmarks/mem_benchmarks/README.md b/benchmarks/mem_benchmarks/README.md
index af1352ad..3854e59f 100644
--- a/benchmarks/mem_benchmarks/README.md
+++ b/benchmarks/mem_benchmarks/README.md
@@ -7,6 +7,6 @@ detection and classification with the small test dataset.
can be used to profile memory useage. Install, and then run
`mprof run --include-children --multiprocess detect_and_classify.py`. It is **very**
important to use these two flags to capture memory usage by the additional
-processes that cellfinder_core uses.
+processes that cellfinder.core uses.
To show the results of the latest profile run, run `mprof plot`.
diff --git a/benchmarks/mem_benchmarks/detect_and_classify.py b/benchmarks/mem_benchmarks/detect_and_classify.py
index ec57d357..c1fd97ab 100644
--- a/benchmarks/mem_benchmarks/detect_and_classify.py
+++ b/benchmarks/mem_benchmarks/detect_and_classify.py
@@ -1,7 +1,7 @@
import dask.array as da
import numpy as np
-from cellfinder_core.main import main
+from cellfinder.core.main import main
voxel_sizes = (5, 2, 2)
diff --git a/benchmarks/mem_benchmarks/filter_2d.py b/benchmarks/mem_benchmarks/filter_2d.py
index fd16172e..1ae24d94 100644
--- a/benchmarks/mem_benchmarks/filter_2d.py
+++ b/benchmarks/mem_benchmarks/filter_2d.py
@@ -1,8 +1,8 @@
import numpy as np
from pyinstrument import Profiler
-from cellfinder_core.detect.filters.plane import TileProcessor
-from cellfinder_core.detect.filters.setup_filters import setup_tile_filtering
+from cellfinder.core.detect.filters.plane import TileProcessor
+from cellfinder.core.detect.filters.setup_filters import setup_tile_filtering
# Use random 16-bit integer data for signal plane
shape = (10000, 10000)
diff --git a/benchmarks/mem_benchmarks/filter_3d.py b/benchmarks/mem_benchmarks/filter_3d.py
index 8afd16a1..5509dd52 100644
--- a/benchmarks/mem_benchmarks/filter_3d.py
+++ b/benchmarks/mem_benchmarks/filter_3d.py
@@ -1,7 +1,7 @@
import numpy as np
from pyinstrument import Profiler
-from cellfinder_core.detect.filters.volume.volume_filter import VolumeFilter
+from cellfinder.core.detect.filters.volume.volume_filter import VolumeFilter
# Use random data for signal data
ball_z_size = 3
diff --git a/src/cellfinder_core/__init__.py b/cellfinder/__init__.py
similarity index 81%
rename from src/cellfinder_core/__init__.py
rename to cellfinder/__init__.py
index 746b2d2b..9971f648 100644
--- a/src/cellfinder_core/__init__.py
+++ b/cellfinder/__init__.py
@@ -1,9 +1,9 @@
from importlib.metadata import PackageNotFoundError, version
try:
- __version__ = version("cellfinder-core")
+ __version__ = version("cellfinder")
except PackageNotFoundError as e:
- raise PackageNotFoundError("cellfinder-core package not installed") from e
+ raise PackageNotFoundError("cellfinder package not installed") from e
# If tensorflow is not present, tools cannot be used.
# Throw an error in this case to prevent invocation of functions.
@@ -22,7 +22,3 @@
__author__ = "Adam Tyson, Christian Niedworok, Charly Rousseau"
__license__ = "BSD-3-Clause"
-
-import logging
-
-logger = logging.getLogger("cellfinder_core")
diff --git a/cellfinder/core/__init__.py b/cellfinder/core/__init__.py
new file mode 100644
index 00000000..213533cc
--- /dev/null
+++ b/cellfinder/core/__init__.py
@@ -0,0 +1,3 @@
+import logging
+
+logger = logging.getLogger("cellfinder.core")
diff --git a/src/cellfinder_core/classify/__init__.py b/cellfinder/core/classify/__init__.py
similarity index 100%
rename from src/cellfinder_core/classify/__init__.py
rename to cellfinder/core/classify/__init__.py
diff --git a/src/cellfinder_core/classify/augment.py b/cellfinder/core/classify/augment.py
similarity index 99%
rename from src/cellfinder_core/classify/augment.py
rename to cellfinder/core/classify/augment.py
index 7951bafa..44a064c9 100644
--- a/src/cellfinder_core/classify/augment.py
+++ b/cellfinder/core/classify/augment.py
@@ -3,7 +3,7 @@
import numpy as np
from scipy.ndimage import rotate, zoom
-from cellfinder_core.tools.tools import (
+from cellfinder.core.tools.tools import (
all_elements_equal,
random_bool,
random_probability,
diff --git a/src/cellfinder_core/classify/classify.py b/cellfinder/core/classify/classify.py
similarity index 92%
rename from src/cellfinder_core/classify/classify.py
rename to cellfinder/core/classify/classify.py
index eb51770e..86b709f5 100644
--- a/src/cellfinder_core/classify/classify.py
+++ b/cellfinder/core/classify/classify.py
@@ -6,10 +6,10 @@
from brainglobe_utils.general.system import get_num_processes
from tensorflow import keras
-from cellfinder_core import logger, types
-from cellfinder_core.classify.cube_generator import CubeGeneratorFromFile
-from cellfinder_core.classify.tools import get_model
-from cellfinder_core.train.train_yml import depth_type, models
+from cellfinder.core import logger, types
+from cellfinder.core.classify.cube_generator import CubeGeneratorFromFile
+from cellfinder.core.classify.tools import get_model
+from cellfinder.core.train.train_yml import depth_type, models
def main(
diff --git a/src/cellfinder_core/classify/cube_generator.py b/cellfinder/core/classify/cube_generator.py
similarity index 99%
rename from src/cellfinder_core/classify/cube_generator.py
rename to cellfinder/core/classify/cube_generator.py
index e617df88..f4c67515 100644
--- a/src/cellfinder_core/classify/cube_generator.py
+++ b/cellfinder/core/classify/cube_generator.py
@@ -10,8 +10,8 @@
from skimage.io import imread
from tensorflow.keras.utils import Sequence
-from cellfinder_core import types
-from cellfinder_core.classify.augment import AugmentationParameters, augment
+from cellfinder.core import types
+from cellfinder.core.classify.augment import AugmentationParameters, augment
# TODO: rename, as now using dask arrays -
# actually should combine to one generator
diff --git a/src/cellfinder_core/classify/resnet.py b/cellfinder/core/classify/resnet.py
similarity index 100%
rename from src/cellfinder_core/classify/resnet.py
rename to cellfinder/core/classify/resnet.py
diff --git a/src/cellfinder_core/classify/tools.py b/cellfinder/core/classify/tools.py
similarity index 96%
rename from src/cellfinder_core/classify/tools.py
rename to cellfinder/core/classify/tools.py
index 020b973d..2d5c44b2 100644
--- a/src/cellfinder_core/classify/tools.py
+++ b/cellfinder/core/classify/tools.py
@@ -5,8 +5,8 @@
import tensorflow as tf
from tensorflow.keras import Model
-from cellfinder_core import logger
-from cellfinder_core.classify.resnet import build_model, layer_type
+from cellfinder.core import logger
+from cellfinder.core.classify.resnet import build_model, layer_type
def get_model(
diff --git a/src/cellfinder_core/config/__init__.py b/cellfinder/core/config/__init__.py
similarity index 100%
rename from src/cellfinder_core/config/__init__.py
rename to cellfinder/core/config/__init__.py
diff --git a/src/cellfinder_core/config/cellfinder.conf b/cellfinder/core/config/cellfinder.conf
similarity index 100%
rename from src/cellfinder_core/config/cellfinder.conf
rename to cellfinder/core/config/cellfinder.conf
diff --git a/src/cellfinder_core/detect/__init__.py b/cellfinder/core/detect/__init__.py
similarity index 100%
rename from src/cellfinder_core/detect/__init__.py
rename to cellfinder/core/detect/__init__.py
diff --git a/src/cellfinder_core/detect/detect.py b/cellfinder/core/detect/detect.py
similarity index 96%
rename from src/cellfinder_core/detect/detect.py
rename to cellfinder/core/detect/detect.py
index a5c63446..94a52239 100644
--- a/src/cellfinder_core/detect/detect.py
+++ b/cellfinder/core/detect/detect.py
@@ -23,10 +23,10 @@
from brainglobe_utils.cells.cells import Cell
from brainglobe_utils.general.system import get_num_processes
-from cellfinder_core import logger, types
-from cellfinder_core.detect.filters.plane import TileProcessor
-from cellfinder_core.detect.filters.setup_filters import setup_tile_filtering
-from cellfinder_core.detect.filters.volume.volume_filter import VolumeFilter
+from cellfinder.core import logger, types
+from cellfinder.core.detect.filters.plane import TileProcessor
+from cellfinder.core.detect.filters.setup_filters import setup_tile_filtering
+from cellfinder.core.detect.filters.volume.volume_filter import VolumeFilter
def calculate_parameters_in_pixels(
diff --git a/src/cellfinder_core/detect/filters/__init__.py b/cellfinder/core/detect/filters/__init__.py
similarity index 100%
rename from src/cellfinder_core/detect/filters/__init__.py
rename to cellfinder/core/detect/filters/__init__.py
diff --git a/src/cellfinder_core/detect/filters/plane/__init__.py b/cellfinder/core/detect/filters/plane/__init__.py
similarity index 100%
rename from src/cellfinder_core/detect/filters/plane/__init__.py
rename to cellfinder/core/detect/filters/plane/__init__.py
diff --git a/src/cellfinder_core/detect/filters/plane/classical_filter.py b/cellfinder/core/detect/filters/plane/classical_filter.py
similarity index 100%
rename from src/cellfinder_core/detect/filters/plane/classical_filter.py
rename to cellfinder/core/detect/filters/plane/classical_filter.py
diff --git a/src/cellfinder_core/detect/filters/plane/plane_filter.py b/cellfinder/core/detect/filters/plane/plane_filter.py
similarity index 94%
rename from src/cellfinder_core/detect/filters/plane/plane_filter.py
rename to cellfinder/core/detect/filters/plane/plane_filter.py
index 3c6a621c..f7e7868e 100644
--- a/src/cellfinder_core/detect/filters/plane/plane_filter.py
+++ b/cellfinder/core/detect/filters/plane/plane_filter.py
@@ -5,9 +5,9 @@
import dask.array as da
import numpy as np
-from cellfinder_core import types
-from cellfinder_core.detect.filters.plane.classical_filter import enhance_peaks
-from cellfinder_core.detect.filters.plane.tile_walker import TileWalker
+from cellfinder.core import types
+from cellfinder.core.detect.filters.plane.classical_filter import enhance_peaks
+from cellfinder.core.detect.filters.plane.tile_walker import TileWalker
@dataclass
diff --git a/src/cellfinder_core/detect/filters/plane/tile_walker.py b/cellfinder/core/detect/filters/plane/tile_walker.py
similarity index 100%
rename from src/cellfinder_core/detect/filters/plane/tile_walker.py
rename to cellfinder/core/detect/filters/plane/tile_walker.py
diff --git a/src/cellfinder_core/detect/filters/setup_filters.py b/cellfinder/core/detect/filters/setup_filters.py
similarity index 90%
rename from src/cellfinder_core/detect/filters/setup_filters.py
rename to cellfinder/core/detect/filters/setup_filters.py
index 4d3de4f7..d68387fc 100644
--- a/src/cellfinder_core/detect/filters/setup_filters.py
+++ b/cellfinder/core/detect/filters/setup_filters.py
@@ -3,11 +3,11 @@
import numpy as np
-from cellfinder_core.detect.filters.volume.ball_filter import BallFilter
-from cellfinder_core.detect.filters.volume.structure_detection import (
+from cellfinder.core.detect.filters.volume.ball_filter import BallFilter
+from cellfinder.core.detect.filters.volume.structure_detection import (
CellDetector,
)
-from cellfinder_core.tools.tools import get_max_possible_value
+from cellfinder.core.tools.tools import get_max_possible_value
def get_ball_filter(
diff --git a/src/cellfinder_core/detect/filters/volume/__init__.py b/cellfinder/core/detect/filters/volume/__init__.py
similarity index 100%
rename from src/cellfinder_core/detect/filters/volume/__init__.py
rename to cellfinder/core/detect/filters/volume/__init__.py
diff --git a/src/cellfinder_core/detect/filters/volume/ball_filter.py b/cellfinder/core/detect/filters/volume/ball_filter.py
similarity index 98%
rename from src/cellfinder_core/detect/filters/volume/ball_filter.py
rename to cellfinder/core/detect/filters/volume/ball_filter.py
index c634b2a2..87dee650 100644
--- a/src/cellfinder_core/detect/filters/volume/ball_filter.py
+++ b/cellfinder/core/detect/filters/volume/ball_filter.py
@@ -1,8 +1,8 @@
import numpy as np
from numba import njit
-from cellfinder_core.tools.array_operations import bin_mean_3d
-from cellfinder_core.tools.geometry import make_sphere
+from cellfinder.core.tools.array_operations import bin_mean_3d
+from cellfinder.core.tools.geometry import make_sphere
DEBUG = False
diff --git a/src/cellfinder_core/detect/filters/volume/structure_detection.py b/cellfinder/core/detect/filters/volume/structure_detection.py
similarity index 100%
rename from src/cellfinder_core/detect/filters/volume/structure_detection.py
rename to cellfinder/core/detect/filters/volume/structure_detection.py
diff --git a/src/cellfinder_core/detect/filters/volume/structure_splitting.py b/cellfinder/core/detect/filters/volume/structure_splitting.py
similarity index 97%
rename from src/cellfinder_core/detect/filters/volume/structure_splitting.py
rename to cellfinder/core/detect/filters/volume/structure_splitting.py
index 1a602c26..4240291a 100644
--- a/src/cellfinder_core/detect/filters/volume/structure_splitting.py
+++ b/cellfinder/core/detect/filters/volume/structure_splitting.py
@@ -2,9 +2,9 @@
import numpy as np
-from cellfinder_core import logger
-from cellfinder_core.detect.filters.volume.ball_filter import BallFilter
-from cellfinder_core.detect.filters.volume.structure_detection import (
+from cellfinder.core import logger
+from cellfinder.core.detect.filters.volume.ball_filter import BallFilter
+from cellfinder.core.detect.filters.volume.structure_detection import (
CellDetector,
get_structure_centre,
)
diff --git a/src/cellfinder_core/detect/filters/volume/volume_filter.py b/cellfinder/core/detect/filters/volume/volume_filter.py
similarity index 96%
rename from src/cellfinder_core/detect/filters/volume/volume_filter.py
rename to cellfinder/core/detect/filters/volume/volume_filter.py
index f92a9585..a3e4d98f 100644
--- a/src/cellfinder_core/detect/filters/volume/volume_filter.py
+++ b/cellfinder/core/detect/filters/volume/volume_filter.py
@@ -9,15 +9,15 @@
from tifffile import tifffile
from tqdm import tqdm
-from cellfinder_core import logger
-from cellfinder_core.detect.filters.setup_filters import (
+from cellfinder.core import logger
+from cellfinder.core.detect.filters.setup_filters import (
get_ball_filter,
get_cell_detector,
)
-from cellfinder_core.detect.filters.volume.structure_detection import (
+from cellfinder.core.detect.filters.volume.structure_detection import (
get_structure_centre,
)
-from cellfinder_core.detect.filters.volume.structure_splitting import (
+from cellfinder.core.detect.filters.volume.structure_splitting import (
StructureSplitException,
split_cells,
)
diff --git a/src/cellfinder_core/download/__init__.py b/cellfinder/core/download/__init__.py
similarity index 100%
rename from src/cellfinder_core/download/__init__.py
rename to cellfinder/core/download/__init__.py
diff --git a/src/cellfinder_core/download/cli.py b/cellfinder/core/download/cli.py
similarity index 94%
rename from src/cellfinder_core/download/cli.py
rename to cellfinder/core/download/cli.py
index bc922e09..e3c6aa46 100644
--- a/src/cellfinder_core/download/cli.py
+++ b/cellfinder/core/download/cli.py
@@ -2,8 +2,8 @@
from argparse import ArgumentDefaultsHelpFormatter, ArgumentParser
from pathlib import Path
-from cellfinder_core.download import models
-from cellfinder_core.download.download import amend_cfg
+from cellfinder.core.download import models
+from cellfinder.core.download.download import amend_cfg
home = Path.home()
DEFAULT_DOWNLOAD_DIRECTORY = home / ".cellfinder"
diff --git a/src/cellfinder_core/download/download.py b/cellfinder/core/download/download.py
similarity index 98%
rename from src/cellfinder_core/download/download.py
rename to cellfinder/core/download/download.py
index 3fe4ecf1..ee242daa 100644
--- a/src/cellfinder_core/download/download.py
+++ b/cellfinder/core/download/download.py
@@ -6,7 +6,7 @@
from brainglobe_utils.general.config import get_config_obj
from brainglobe_utils.general.system import disk_free_gb
-from cellfinder_core.tools.source_files import (
+from cellfinder.core.tools.source_files import (
source_config_cellfinder,
source_custom_config_cellfinder,
)
diff --git a/src/cellfinder_core/download/models.py b/cellfinder/core/download/models.py
similarity index 93%
rename from src/cellfinder_core/download/models.py
rename to cellfinder/core/download/models.py
index b3905b54..dbb0f3cb 100644
--- a/src/cellfinder_core/download/models.py
+++ b/cellfinder/core/download/models.py
@@ -2,8 +2,8 @@
from pathlib import Path
from typing import Literal
-from cellfinder_core import logger
-from cellfinder_core.download.download import download
+from cellfinder.core import logger
+from cellfinder.core.download.download import download
model_weight_urls = {
"resnet50_tv": "https://gin.g-node.org/cellfinder/models/raw/"
diff --git a/src/cellfinder_core/main.py b/cellfinder/core/main.py
similarity index 92%
rename from src/cellfinder_core/main.py
rename to cellfinder/core/main.py
index afbbcf1c..4ad395e2 100644
--- a/src/cellfinder_core/main.py
+++ b/cellfinder/core/main.py
@@ -8,9 +8,9 @@
import numpy as np
from brainglobe_utils.general.logging import suppress_specific_logs
-from cellfinder_core import logger
-from cellfinder_core.download.models import model_type
-from cellfinder_core.train.train_yml import depth_type
+from cellfinder.core import logger
+from cellfinder.core.download.models import model_type
+from cellfinder.core.train.train_yml import depth_type
tf_suppress_log_messages = [
"multiprocessing can interact badly with TensorFlow"
@@ -60,9 +60,9 @@ def main(
"""
suppress_tf_logging(tf_suppress_log_messages)
- from cellfinder_core.classify import classify
- from cellfinder_core.detect import detect
- from cellfinder_core.tools import prep
+ from cellfinder.core.classify import classify
+ from cellfinder.core.detect import detect
+ from cellfinder.core.tools import prep
logger.info("Detecting cell candidates")
diff --git a/src/cellfinder_core/tools/IO.py b/cellfinder/core/tools/IO.py
similarity index 100%
rename from src/cellfinder_core/tools/IO.py
rename to cellfinder/core/tools/IO.py
diff --git a/src/cellfinder_core/tools/__init__.py b/cellfinder/core/tools/__init__.py
similarity index 100%
rename from src/cellfinder_core/tools/__init__.py
rename to cellfinder/core/tools/__init__.py
diff --git a/src/cellfinder_core/tools/array_operations.py b/cellfinder/core/tools/array_operations.py
similarity index 100%
rename from src/cellfinder_core/tools/array_operations.py
rename to cellfinder/core/tools/array_operations.py
diff --git a/src/cellfinder_core/tools/geometry.py b/cellfinder/core/tools/geometry.py
similarity index 100%
rename from src/cellfinder_core/tools/geometry.py
rename to cellfinder/core/tools/geometry.py
diff --git a/src/cellfinder_core/tools/image_processing.py b/cellfinder/core/tools/image_processing.py
similarity index 100%
rename from src/cellfinder_core/tools/image_processing.py
rename to cellfinder/core/tools/image_processing.py
diff --git a/src/cellfinder_core/tools/prep.py b/cellfinder/core/tools/prep.py
similarity index 91%
rename from src/cellfinder_core/tools/prep.py
rename to cellfinder/core/tools/prep.py
index d9637144..dc2eda54 100644
--- a/src/cellfinder_core/tools/prep.py
+++ b/cellfinder/core/tools/prep.py
@@ -10,11 +10,11 @@
from brainglobe_utils.general.config import get_config_obj
from brainglobe_utils.general.system import get_num_processes
-import cellfinder_core.tools.tf as tf_tools
-from cellfinder_core import logger
-from cellfinder_core.download import models as model_download
-from cellfinder_core.download.download import amend_cfg
-from cellfinder_core.tools.source_files import source_custom_config_cellfinder
+import cellfinder.core.tools.tf as tf_tools
+from cellfinder.core import logger
+from cellfinder.core.download import models as model_download
+from cellfinder.core.download.download import amend_cfg
+from cellfinder.core.tools.source_files import source_custom_config_cellfinder
home = Path.home()
DEFAULT_INSTALL_PATH = home / ".cellfinder"
diff --git a/src/cellfinder_core/tools/source_files.py b/cellfinder/core/tools/source_files.py
similarity index 100%
rename from src/cellfinder_core/tools/source_files.py
rename to cellfinder/core/tools/source_files.py
diff --git a/src/cellfinder_core/tools/system.py b/cellfinder/core/tools/system.py
similarity index 100%
rename from src/cellfinder_core/tools/system.py
rename to cellfinder/core/tools/system.py
diff --git a/src/cellfinder_core/tools/tf.py b/cellfinder/core/tools/tf.py
similarity index 97%
rename from src/cellfinder_core/tools/tf.py
rename to cellfinder/core/tools/tf.py
index 60ea30c3..778aa78f 100644
--- a/src/cellfinder_core/tools/tf.py
+++ b/cellfinder/core/tools/tf.py
@@ -1,6 +1,6 @@
import tensorflow as tf
-from cellfinder_core import logger
+from cellfinder.core import logger
def allow_gpu_memory_growth():
diff --git a/src/cellfinder_core/tools/tiff.py b/cellfinder/core/tools/tiff.py
similarity index 100%
rename from src/cellfinder_core/tools/tiff.py
rename to cellfinder/core/tools/tiff.py
diff --git a/src/cellfinder_core/tools/tools.py b/cellfinder/core/tools/tools.py
similarity index 100%
rename from src/cellfinder_core/tools/tools.py
rename to cellfinder/core/tools/tools.py
diff --git a/src/cellfinder_core/train/__init__.py b/cellfinder/core/train/__init__.py
similarity index 100%
rename from src/cellfinder_core/train/__init__.py
rename to cellfinder/core/train/__init__.py
diff --git a/src/cellfinder_core/train/train_yml.py b/cellfinder/core/train/train_yml.py
similarity index 95%
rename from src/cellfinder_core/train/train_yml.py
rename to cellfinder/core/train/train_yml.py
index 47f3ec53..5596dcff 100644
--- a/src/cellfinder_core/train/train_yml.py
+++ b/cellfinder/core/train/train_yml.py
@@ -28,10 +28,10 @@
from fancylog import fancylog
from sklearn.model_selection import train_test_split
-import cellfinder_core as program_for_log
-from cellfinder_core import logger
-from cellfinder_core.classify.resnet import layer_type
-from cellfinder_core.tools.prep import DEFAULT_INSTALL_PATH
+import cellfinder.core as program_for_log
+from cellfinder.core import logger
+from cellfinder.core.classify.resnet import layer_type
+from cellfinder.core.tools.prep import DEFAULT_INSTALL_PATH
tf_suppress_log_messages = [
"sample_weight modes were coerced from",
@@ -111,7 +111,7 @@ def misc_parse(parser):
def training_parse():
- from cellfinder_core.download.cli import (
+ from cellfinder.core.download.cli import (
download_directory_parser,
model_parser,
)
@@ -238,7 +238,7 @@ def parse_yaml(yaml_files, section="data"):
def get_tiff_files(yaml_contents):
- from cellfinder_core.tools.tiff import TiffDir, TiffList
+ from cellfinder.core.tools.tiff import TiffDir, TiffList
tiff_lists = []
for d in yaml_contents:
@@ -320,7 +320,7 @@ def run(
save_progress=False,
epochs=100,
):
- from cellfinder_core.main import suppress_tf_logging
+ from cellfinder.core.main import suppress_tf_logging
suppress_tf_logging(tf_suppress_log_messages)
@@ -330,9 +330,9 @@ def run(
TensorBoard,
)
- from cellfinder_core.classify.cube_generator import CubeGeneratorFromDisk
- from cellfinder_core.classify.tools import get_model, make_lists
- from cellfinder_core.tools.prep import prep_training
+ from cellfinder.core.classify.cube_generator import CubeGeneratorFromDisk
+ from cellfinder.core.classify.tools import get_model, make_lists
+ from cellfinder.core.tools.prep import prep_training
start_time = datetime.now()
diff --git a/src/cellfinder_core/types.py b/cellfinder/core/types.py
similarity index 100%
rename from src/cellfinder_core/types.py
rename to cellfinder/core/types.py
diff --git a/cellfinder/napari/__init__.py b/cellfinder/napari/__init__.py
new file mode 100644
index 00000000..0d6ef0a4
--- /dev/null
+++ b/cellfinder/napari/__init__.py
@@ -0,0 +1,3 @@
+__version__ = "0.0.20"
+__author__ = "Adam Tyson"
+__license__ = "GPL-3.0"
diff --git a/cellfinder/napari/curation.py b/cellfinder/napari/curation.py
new file mode 100644
index 00000000..bf6258d1
--- /dev/null
+++ b/cellfinder/napari/curation.py
@@ -0,0 +1,591 @@
+from pathlib import Path
+from typing import List, Optional, Tuple
+
+import napari
+import numpy as np
+import tifffile
+from brainglobe_napari_io.cellfinder.utils import convert_layer_to_cells
+from brainglobe_utils.cells.cells import Cell
+from brainglobe_utils.IO.yaml import save_yaml
+from magicgui.widgets import ProgressBar
+from napari.qt.threading import thread_worker
+from napari.utils.notifications import show_info
+from qtpy import QtCore
+from qtpy.QtWidgets import (
+ QComboBox,
+ QFileDialog,
+ QGridLayout,
+ QGroupBox,
+ QLabel,
+ QWidget,
+)
+
+from .utils import add_button, add_combobox, display_question
+
+# Constants used throughout
+WINDOW_HEIGHT = 750
+WINDOW_WIDTH = 1500
+COLUMN_WIDTH = 150
+
+
+class CurationWidget(QWidget):
+ def __init__(
+ self,
+ viewer: napari.viewer.Viewer,
+ cube_depth: int = 20,
+ cube_width: int = 50,
+ cube_height: int = 50,
+ network_voxel_sizes: Tuple[int, int, int] = (5, 1, 1),
+ n_free_cpus: int = 2,
+ save_empty_cubes: bool = False,
+ max_ram=None,
+ ):
+ super(CurationWidget, self).__init__()
+
+ self.non_cells_to_extract = None
+ self.cells_to_extract = None
+
+ self.cube_depth = cube_depth
+ self.cube_width = cube_width
+ self.cube_height = cube_height
+ self.network_voxel_sizes = network_voxel_sizes
+ self.n_free_cpus = n_free_cpus
+ self.save_empty_cubes = save_empty_cubes
+ self.max_ram = max_ram
+ self.voxel_sizes = [5, 2, 2]
+ self.batch_size = 32
+ self.viewer = viewer
+
+ self.signal_layer = None
+ self.background_layer = None
+ self.training_data_cell_layer = None
+ self.training_data_non_cell_layer = None
+
+ self.image_layer_names = self._get_layer_names()
+ self.point_layer_names = self._get_layer_names(
+ layer_type=napari.layers.Points
+ )
+
+ self.output_directory: Optional[Path] = None
+
+ self.setup_main_layout()
+
+ @self.viewer.layers.events.connect
+ def update_layer_list(v: napari.viewer.Viewer):
+ """
+ Update internal list of layers whenever the napari layers list
+ is updated.
+ """
+ self.image_layer_names = self._get_layer_names()
+ self.point_layer_names = self._get_layer_names(
+ layer_type=napari.layers.Points
+ )
+ self._update_combobox_options(
+ self.signal_image_choice, self.image_layer_names
+ )
+ self._update_combobox_options(
+ self.background_image_choice, self.image_layer_names
+ )
+ self._update_combobox_options(
+ self.training_data_cell_choice, self.point_layer_names
+ )
+ self._update_combobox_options(
+ self.training_data_non_cell_choice, self.point_layer_names
+ )
+
+ @staticmethod
+ def _update_combobox_options(combobox: QComboBox, options_list: List[str]):
+ original_text = combobox.currentText()
+ combobox.clear()
+ combobox.addItems(options_list)
+ combobox.setCurrentText(original_text)
+
+ def _get_layer_names(
+ self,
+ layer_type: napari.layers.Layer = napari.layers.Image,
+ default: str = "",
+ ) -> List[str]:
+ """
+ Get list of layer names of a given layer type.
+ """
+ layer_names = [
+ layer.name
+ for layer in self.viewer.layers
+ if type(layer) == layer_type
+ ]
+
+ if layer_names:
+ return [default] + layer_names
+ else:
+ return [default]
+
+ def setup_main_layout(self):
+ """
+ Construct main layout of widget.
+ """
+ self.layout = QGridLayout()
+ self.layout.setContentsMargins(10, 10, 10, 10)
+ self.layout.setAlignment(QtCore.Qt.AlignTop)
+ self.layout.setSpacing(4)
+
+ self.add_loading_panel(1)
+
+ self.status_label = QLabel()
+ row, col = 7, 0
+ self.layout.addWidget(self.status_label, row, col)
+ self.update_status_label("Ready")
+
+ self.progress_bar = ProgressBar()
+ row, col = 8, 0
+ self.layout.addWidget(self.progress_bar.native, row, col)
+
+ self.setLayout(self.layout)
+
+ def add_loading_panel(self, row: int, column: int = 0):
+ self.load_data_panel = QGroupBox("Load data")
+ self.load_data_layout = QGridLayout()
+ self.load_data_layout.setSpacing(15)
+ self.load_data_layout.setContentsMargins(10, 10, 10, 10)
+ self.load_data_layout.setAlignment(QtCore.Qt.AlignBottom)
+
+ self.signal_image_choice, _ = add_combobox(
+ self.load_data_layout,
+ "Signal image",
+ self.image_layer_names,
+ 1,
+ callback=self.set_signal_image,
+ )
+ self.background_image_choice, _ = add_combobox(
+ self.load_data_layout,
+ "Background image",
+ self.image_layer_names,
+ 2,
+ callback=self.set_background_image,
+ )
+ self.training_data_cell_choice, _ = add_combobox(
+ self.load_data_layout,
+ "Training data (cells)",
+ self.point_layer_names,
+ 3,
+ callback=self.set_training_data_cell,
+ )
+ self.training_data_non_cell_choice, _ = add_combobox(
+ self.load_data_layout,
+ "Training_data (non_cells)",
+ self.point_layer_names,
+ 4,
+ callback=self.set_training_data_non_cell,
+ )
+ self.mark_as_cell_button = add_button(
+ "Mark as cell(s)",
+ self.load_data_layout,
+ self.mark_as_cell,
+ 5,
+ )
+ self.mark_as_non_cell_button = add_button(
+ "Mark as non cell(s)",
+ self.load_data_layout,
+ self.mark_as_non_cell,
+ 5,
+ column=1,
+ )
+ self.add_training_data_button = add_button(
+ "Add training data layers",
+ self.load_data_layout,
+ self.add_training_data,
+ 6,
+ )
+ self.save_training_data_button = add_button(
+ "Save training data",
+ self.load_data_layout,
+ self.save_training_data,
+ 6,
+ column=1,
+ )
+ self.load_data_layout.setColumnMinimumWidth(0, COLUMN_WIDTH)
+ self.load_data_panel.setLayout(self.load_data_layout)
+ self.load_data_panel.setVisible(True)
+ self.layout.addWidget(self.load_data_panel, row, column, 1, 1)
+
+ def set_signal_image(self):
+ """
+ Set signal layer from current signal text box selection.
+ """
+ if self.signal_image_choice.currentText() != "":
+ self.signal_layer = self.viewer.layers[
+ self.signal_image_choice.currentText()
+ ]
+
+ def set_background_image(self):
+ """
+ Set background layer from current background text box selection.
+ """
+ if self.background_image_choice.currentText() != "":
+ self.background_layer = self.viewer.layers[
+ self.background_image_choice.currentText()
+ ]
+
+ def set_training_data_cell(self):
+ """
+ Set cell training data from current training data text box selection.
+ """
+ if self.training_data_cell_choice.currentText() != "":
+ self.training_data_cell_layer = self.viewer.layers[
+ self.training_data_cell_choice.currentText()
+ ]
+ self.training_data_cell_layer.metadata["point_type"] = Cell.CELL
+ self.training_data_cell_layer.metadata["training_data"] = True
+
+ def set_training_data_non_cell(self):
+ """
+ Set non-cell training data from current training data text box
+ selection.
+ """
+ if self.training_data_non_cell_choice.currentText() != "":
+ self.training_data_non_cell_layer = self.viewer.layers[
+ self.training_data_non_cell_choice.currentText()
+ ]
+ self.training_data_non_cell_layer.metadata[
+ "point_type"
+ ] = Cell.UNKNOWN
+ self.training_data_non_cell_layer.metadata["training_data"] = True
+
+ def add_training_data(self):
+ cell_name = "Training data (cells)"
+ non_cell_name = "Training data (non cells)"
+
+ overwrite = False
+ if self.training_data_cell_layer or self.training_data_non_cell_layer:
+ overwrite = display_question(
+ self,
+ "Training data layers exist",
+ "Training data layers already exist, "
+ "overwrite with empty layers?.",
+ )
+ else:
+ if self.training_data_cell_layer:
+ self.training_data_cell_layer.remove()
+ self._add_training_data_layers(cell_name, non_cell_name)
+
+ if overwrite:
+ try:
+ self.viewer.layers.remove(cell_name)
+ self.viewer.layers.remove(non_cell_name)
+ except ValueError:
+ pass
+
+ self._add_training_data_layers(cell_name, non_cell_name)
+
+ def _add_training_data_layers(self, cell_name: str, non_cell_name: str):
+ self.training_data_cell_layer = self.viewer.add_points(
+ None,
+ ndim=3,
+ symbol="ring",
+ n_dimensional=True,
+ size=15,
+ opacity=0.6,
+ face_color="lightgoldenrodyellow",
+ name=cell_name,
+ metadata=dict(point_type=Cell.CELL, training_data=True),
+ )
+ self.training_data_cell_choice.setCurrentText(cell_name)
+
+ self.training_data_non_cell_layer = self.viewer.add_points(
+ None,
+ ndim=3,
+ symbol="ring",
+ n_dimensional=True,
+ size=15,
+ opacity=0.6,
+ face_color="lightskyblue",
+ name=non_cell_name,
+ metadata=dict(point_type=Cell.UNKNOWN, training_data=True),
+ )
+ self.training_data_non_cell_choice.setCurrentText(non_cell_name)
+
+ def mark_as_cell(self):
+ self.mark_point_as_type("cell")
+
+ def mark_as_non_cell(self):
+ self.mark_point_as_type("non-cell")
+
+ def mark_point_as_type(self, point_type: str):
+ if not (
+ self.training_data_cell_layer and self.training_data_non_cell_layer
+ ):
+ show_info(
+ "No training data layers have been chosen. "
+ "Please add training data layers. ",
+ )
+ return
+
+ if len(self.viewer.layers.selection) == 1:
+ layer = list(self.viewer.layers.selection)[0]
+ if type(layer) == napari.layers.Points:
+ if len(layer.data) > 0:
+ if point_type == "cell":
+ destination_layer = self.training_data_cell_layer
+ else:
+ destination_layer = self.training_data_non_cell_layer
+ show_info(
+ f"Adding {len(layer.selected_data)} "
+ f"points to layer: {destination_layer.name}"
+ )
+
+ for selected_point in layer.selected_data:
+ destination_layer.data = np.vstack(
+ (
+ destination_layer.data,
+ layer.data[selected_point],
+ )
+ )
+
+ else:
+ show_info(
+ "No points are selected in the current layer. "
+ "Please select some points.",
+ )
+
+ else:
+ show_info(
+ "This is not a points layer. "
+ "Please choose a points layer, and select some points.",
+ )
+ elif len(self.viewer.layers.selected) == 0:
+ show_info(
+ "No layers are selected. "
+ "Please choose a single points layer, and select some points.",
+ )
+ else:
+ show_info(
+ "More than one layer is selected. "
+ "Please choose a single points layer, and select some points.",
+ )
+
+ def save_training_data(
+ self, *, block: bool = False, prompt_for_directory: bool = True
+ ) -> None:
+ """
+ Parameters
+ ----------
+ block :
+ If `True` block execution until all cubes are saved.
+ prompt_for_directory :
+ If `True` show a file dialog for the user to select a directory.
+ """
+ if self.is_data_extractable():
+ if prompt_for_directory:
+ self.get_output_directory()
+ if self.output_directory is not None:
+ self.__extract_cubes(block=block)
+ self.__save_yaml_file()
+ show_info("Done")
+
+ self.update_status_label("Ready")
+
+ def __extract_cubes(self, *, block=False):
+ """
+ Parameters
+ ----------
+ block :
+ If `True` block execution until all cubes are saved.
+ """
+ self.update_status_label("Extracting cubes")
+ self.convert_layers_to_cells()
+
+ if block:
+ cubes = self.extract_cubes()
+ while True:
+ try:
+ next(cubes)
+ except StopIteration:
+ break
+ else:
+
+ @thread_worker(connect={"yielded": self.update_progress})
+ def extract_cubes():
+ yield from self.extract_cubes()
+
+ extract_cubes()
+
+ def is_data_extractable(self) -> bool:
+ if (
+ self.check_training_data_exists()
+ and self.check_image_data_for_extraction()
+ ):
+ return True
+ else:
+ return False
+
+ def check_image_data_for_extraction(self) -> bool:
+ if self.signal_layer and self.background_layer:
+ if (
+ self.signal_layer.data.shape
+ == self.background_layer.data.shape
+ ):
+ return True
+ else:
+ show_info(
+ "Please ensure both signal and background images are the "
+ "same size and shape.",
+ )
+ return False
+
+ else:
+ show_info(
+ "Please ensure both signal and background images are loaded "
+ "into napari, and selected in the sidebar. ",
+ )
+ return False
+
+ def check_training_data_exists(self) -> bool:
+ if not (
+ self.training_data_cell_layer or self.training_data_non_cell_layer
+ ):
+ show_info(
+ "No training data layers have been added. "
+ "Please add a layer and annotate some points.",
+ )
+ return False
+ else:
+ if (
+ len(self.training_data_cell_layer.data) > 0
+ or len(self.training_data_non_cell_layer.data) > 0
+ ):
+ return True
+ else:
+ show_info(
+ "No training data points have been added. "
+ "Please annotate some points.",
+ )
+ return False
+
+ def get_output_directory(self):
+ """
+ Shows file dialog to choose output directory
+ """
+ self.update_status_label("Setting output directory...")
+ options = QFileDialog.Options()
+ options |= QFileDialog.DontUseNativeDialog
+ self.output_directory = QFileDialog.getExistingDirectory(
+ self,
+ "Select output directory",
+ options=options,
+ )
+ if self.output_directory != "":
+ self.output_directory = Path(self.output_directory)
+ else:
+ self.output_directory = None
+
+ def convert_layers_to_cells(self):
+ self.cells_to_extract = convert_layer_to_cells(
+ self.training_data_cell_layer.data
+ )
+ self.non_cells_to_extract = convert_layer_to_cells(
+ self.training_data_non_cell_layer.data, cells=False
+ )
+
+ self.cells_to_extract = list(set(self.cells_to_extract))
+ self.non_cells_to_extract = list(set(self.non_cells_to_extract))
+
+ def __save_yaml_file(self):
+ # TODO: implement this in a portable way
+ yaml_filename = self.output_directory / "training.yml"
+ yaml_section = [
+ {
+ "cube_dir": str(self.output_directory / "cells"),
+ "cell_def": "",
+ "type": "cell",
+ "signal_channel": 0,
+ "bg_channel": 1,
+ },
+ {
+ "cube_dir": str(self.output_directory / "non_cells"),
+ "cell_def": "",
+ "type": "no_cell",
+ "signal_channel": 0,
+ "bg_channel": 1,
+ },
+ ]
+
+ yaml_contents = {"data": yaml_section}
+ save_yaml(yaml_contents, yaml_filename)
+
+ def update_progress(self, attributes: dict):
+ """
+ Update progress bar with ``attributes``.
+ """
+ for attr in attributes:
+ self.progress_bar.__setattr__(attr, attributes[attr])
+
+ def update_status_label(self, label: str):
+ self.status_label.setText(label)
+
+ def extract_cubes(self):
+ """
+ Yields
+ ------
+ dict
+ Attributes used to update a progress bar. The keys can be any of
+ the properties of `magicgui.widgets.ProgressBar`.
+ """
+ from cellfinder.core.classify.cube_generator import (
+ CubeGeneratorFromFile,
+ )
+
+ to_extract = {
+ "cells": self.cells_to_extract,
+ "non_cells": self.non_cells_to_extract,
+ }
+
+ for cell_type, cell_list in to_extract.items():
+ cell_type_output_directory = self.output_directory / cell_type
+ cell_type_output_directory.mkdir(exist_ok=True, parents=True)
+ self.update_status_label(f"Saving {cell_type}...")
+
+ cube_generator = CubeGeneratorFromFile(
+ cell_list,
+ self.signal_layer.data,
+ self.background_layer.data,
+ self.voxel_sizes,
+ self.network_voxel_sizes,
+ batch_size=self.batch_size,
+ cube_width=self.cube_width,
+ cube_height=self.cube_height,
+ cube_depth=self.cube_depth,
+ extract=True,
+ )
+ # Set up progress bar
+ yield {
+ "value": 0,
+ "min": 0,
+ "max": len(cube_generator),
+ }
+
+ for i, (image_batch, batch_info) in enumerate(cube_generator):
+ image_batch = image_batch.astype(np.int16)
+
+ for point, point_info in zip(image_batch, batch_info):
+ point = np.moveaxis(point, 2, 0)
+ for channel in range(point.shape[-1]):
+ save_cube(
+ point,
+ point_info,
+ channel,
+ cell_type_output_directory,
+ )
+
+ # Update progress bar
+ yield {"value": i + 1}
+
+ self.update_status_label("Finished saving cubes")
+
+
+def save_cube(
+ array: np.ndarray, point_info: dict, channel: int, output_directory: Path
+):
+ filename = (
+ f"pCellz{point_info['z']}y{point_info['y']}"
+ f"x{point_info['x']}Ch{channel}.tif"
+ )
+ tifffile.imwrite(output_directory / filename, array[:, :, :, channel])
diff --git a/cellfinder/napari/detect/__init__.py b/cellfinder/napari/detect/__init__.py
new file mode 100644
index 00000000..616762e4
--- /dev/null
+++ b/cellfinder/napari/detect/__init__.py
@@ -0,0 +1 @@
+from .detect import detect_widget
diff --git a/cellfinder/napari/detect/detect.py b/cellfinder/napari/detect/detect.py
new file mode 100644
index 00000000..396f90b6
--- /dev/null
+++ b/cellfinder/napari/detect/detect.py
@@ -0,0 +1,233 @@
+from math import ceil
+from pathlib import Path
+from typing import Optional
+
+import napari
+from magicgui import magicgui
+from magicgui.widgets import FunctionGui, ProgressBar
+from napari.utils.notifications import show_info
+from qtpy.QtWidgets import QScrollArea
+
+from cellfinder.core.classify.cube_generator import get_cube_depth_min_max
+from cellfinder.napari.utils import (
+ add_layers,
+ header_label_widget,
+ html_label_widget,
+ widget_header,
+)
+
+from .detect_containers import (
+ ClassificationInputs,
+ DataInputs,
+ DetectionInputs,
+ MiscInputs,
+)
+from .thread_worker import Worker
+
+NETWORK_VOXEL_SIZES = [5, 1, 1]
+CUBE_WIDTH = 50
+CUBE_HEIGHT = 20
+CUBE_DEPTH = 20
+
+# If using ROI, how many extra planes to analyse
+MIN_PLANES_ANALYSE = 0
+
+
+def detect_widget() -> FunctionGui:
+ """
+ Create a detection plugin GUI.
+ """
+ progress_bar = ProgressBar()
+
+ @magicgui(
+ header=header_label_widget,
+ detection_label=html_label_widget("Cell detection", tag="h3"),
+ **DataInputs.widget_representation(),
+ **DetectionInputs.widget_representation(),
+ **ClassificationInputs.widget_representation(),
+ **MiscInputs.widget_representation(),
+ call_button=True,
+ persist=True,
+ reset_button=dict(widget_type="PushButton", text="Reset defaults"),
+ scrollable=True,
+ )
+ def widget(
+ header,
+ detection_label,
+ data_options,
+ viewer: napari.Viewer,
+ signal_image: napari.layers.Image,
+ background_image: napari.layers.Image,
+ voxel_size_z: float,
+ voxel_size_y: float,
+ voxel_size_x: float,
+ detection_options,
+ soma_diameter: float,
+ ball_xy_size: float,
+ ball_z_size: float,
+ ball_overlap_fraction: float,
+ log_sigma_size: float,
+ n_sds_above_mean_thresh: int,
+ soma_spread_factor: float,
+ max_cluster_size: int,
+ classification_options,
+ trained_model: Optional[Path],
+ use_pre_trained_weights: bool,
+ misc_options,
+ start_plane: int,
+ end_plane: int,
+ n_free_cpus: int,
+ analyse_local: bool,
+ debug: bool,
+ reset_button,
+ ) -> None:
+ """
+ Run detection and classification.
+
+ Parameters
+ ----------
+ signal_image : napari.layers.Image
+ Image layer containing the labelled cells
+ background_image : napari.layers.Image
+ Image layer without labelled cells
+ voxel_size_z : float
+ Size of your voxels in the axial dimension
+ voxel_size_y : float
+ Size of your voxels in the y direction (top to bottom)
+ voxel_size_x : float
+ Size of your voxels in the x direction (left to right)
+ soma_diameter : float
+ The expected in-plane soma diameter (microns)
+ ball_xy_size : float
+ Elliptical morphological in-plane filter size (microns)
+ ball_z_size : float
+ Elliptical morphological axial filter size (microns)
+ ball_overlap_fraction : float
+ Fraction of the morphological filter needed to be filled
+ to retain a voxel
+ log_sigma_size : float
+ Laplacian of Gaussian filter width (as a fraction of soma diameter)
+ n_sds_above_mean_thresh : int
+ Cell intensity threshold (as a multiple of noise above the mean)
+ soma_spread_factor : float
+ Cell spread factor (for splitting up cell clusters)
+ max_cluster_size : int
+ Largest putative cell cluster (in cubic um) where splitting
+ should be attempted
+ use_pre_trained_weights : bool
+ Select to use pre-trained model weights
+ trained_model : Optional[Path]
+ Trained model file path (home directory (default) -> pretrained
+ weights)
+ start_plane : int
+ First plane to process (to process a subset of the data)
+ end_plane : int
+ Last plane to process (to process a subset of the data)
+ n_free_cpus : int
+ How many CPU cores to leave free
+ analyse_local : bool
+ Only analyse planes around the current position
+ debug : bool
+ Increase logging
+ reset_button :
+ Reset parameters to default
+ """
+ if signal_image is None or background_image is None:
+ show_info("Both signal and background images must be specified.")
+ return
+ data_inputs = DataInputs(
+ signal_image.data,
+ background_image.data,
+ voxel_size_z,
+ voxel_size_y,
+ voxel_size_x,
+ )
+
+ detection_inputs = DetectionInputs(
+ soma_diameter,
+ ball_xy_size,
+ ball_z_size,
+ ball_overlap_fraction,
+ log_sigma_size,
+ n_sds_above_mean_thresh,
+ soma_spread_factor,
+ max_cluster_size,
+ )
+
+ if use_pre_trained_weights:
+ trained_model = None
+ classification_inputs = ClassificationInputs(
+ use_pre_trained_weights, trained_model
+ )
+
+ end_plane = len(signal_image.data) if end_plane == 0 else end_plane
+
+ if analyse_local:
+ current_plane = viewer.dims.current_step[0]
+
+ # so a reasonable number of cells in the plane are detected
+ planes_needed = MIN_PLANES_ANALYSE + int(
+ ceil((CUBE_DEPTH * NETWORK_VOXEL_SIZES[0]) / voxel_size_z)
+ )
+
+ start_plane, end_plane = get_cube_depth_min_max(
+ current_plane, planes_needed
+ )
+ start_plane = max(0, start_plane)
+ end_plane = min(len(signal_image.data), end_plane)
+
+ misc_inputs = MiscInputs(
+ start_plane, end_plane, n_free_cpus, analyse_local, debug
+ )
+
+ worker = Worker(
+ data_inputs,
+ detection_inputs,
+ classification_inputs,
+ misc_inputs,
+ )
+ worker.returned.connect(
+ lambda points: add_layers(points, viewer=viewer)
+ )
+
+ # Make sure if the worker emits an error, it is propagated to this
+ # thread
+ def reraise(e):
+ raise Exception from e
+
+ worker.errored.connect(reraise)
+
+ def update_progress_bar(label: str, max: int, value: int):
+ progress_bar.label = label
+ progress_bar.max = max
+ progress_bar.value = value
+
+ worker.update_progress_bar.connect(update_progress_bar)
+ worker.start()
+
+ widget.header.value = widget_header
+ widget.header.native.setOpenExternalLinks(True)
+
+ @widget.reset_button.changed.connect
+ def restore_defaults():
+ """
+ Restore default widget values.
+ """
+ defaults = {
+ **DataInputs.defaults(),
+ **DetectionInputs.defaults(),
+ **ClassificationInputs.defaults(),
+ **MiscInputs.defaults(),
+ }
+ for name, value in defaults.items():
+ if value is not None: # ignore fields with no default
+ getattr(widget, name).value = value
+
+ # Insert progress bar before the run and reset buttons
+ widget.insert(-3, progress_bar)
+
+ scroll = QScrollArea()
+ scroll.setWidget(widget._widget._qwidget)
+ widget._widget._qwidget = scroll
+
+ return widget
diff --git a/cellfinder/napari/detect/detect_containers.py b/cellfinder/napari/detect/detect_containers.py
new file mode 100644
index 00000000..824a2a0b
--- /dev/null
+++ b/cellfinder/napari/detect/detect_containers.py
@@ -0,0 +1,156 @@
+from dataclasses import dataclass
+from pathlib import Path
+from typing import Optional
+
+import numpy
+
+from cellfinder.napari.input_container import InputContainer
+from cellfinder.napari.utils import html_label_widget
+
+
+@dataclass
+class DataInputs(InputContainer):
+ """Container for image-related ("Data") inputs."""
+
+ signal_array: numpy.ndarray = None
+ background_array: numpy.ndarray = None
+ voxel_size_z: float = 5
+ voxel_size_y: float = 2
+ voxel_size_x: float = 2
+
+ def as_core_arguments(self) -> dict:
+ """
+ Passes voxel size data as one tuple instead of 3 individual floats
+ """
+ data_input_dict = super().as_core_arguments()
+ data_input_dict["voxel_sizes"] = (
+ self.voxel_size_z,
+ self.voxel_size_y,
+ self.voxel_size_x,
+ )
+ # del operator doesn't affect self, because asdict creates a copy of
+ # fields.
+ del data_input_dict["voxel_size_z"]
+ del data_input_dict["voxel_size_y"]
+ del data_input_dict["voxel_size_x"]
+ return data_input_dict
+
+ @property
+ def nplanes(self):
+ return len(self.signal_array)
+
+ @classmethod
+ def widget_representation(cls) -> dict:
+ return dict(
+ data_options=html_label_widget("Data:"),
+ voxel_size_z=cls._custom_widget(
+ "voxel_size_z", custom_label="Voxel size (z)"
+ ),
+ voxel_size_y=cls._custom_widget(
+ "voxel_size_y", custom_label="Voxel size (y)"
+ ),
+ voxel_size_x=cls._custom_widget(
+ "voxel_size_x", custom_label="Voxel size (x)"
+ ),
+ )
+
+
+@dataclass
+class DetectionInputs(InputContainer):
+ """Container for cell candidate detection inputs."""
+
+ soma_diameter: float = 16.0
+ ball_xy_size: float = 6
+ ball_z_size: float = 15
+ ball_overlap_fraction: float = 0.6
+ log_sigma_size: float = 0.2
+ n_sds_above_mean_thresh: int = 10
+ soma_spread_factor: float = 1.4
+ max_cluster_size: int = 100000
+
+ def as_core_arguments(self) -> dict:
+ return super().as_core_arguments()
+
+ @classmethod
+ def widget_representation(cls) -> dict:
+ return dict(
+ detection_options=html_label_widget("Detection:"),
+ soma_diameter=cls._custom_widget("soma_diameter"),
+ ball_xy_size=cls._custom_widget(
+ "ball_xy_size", custom_label="Ball filter (xy)"
+ ),
+ ball_z_size=cls._custom_widget(
+ "ball_z_size", custom_label="Ball filter (z)"
+ ),
+ ball_overlap_fraction=cls._custom_widget(
+ "ball_overlap_fraction", custom_label="Ball overlap"
+ ),
+ log_sigma_size=cls._custom_widget(
+ "log_sigma_size", custom_label="Filter width"
+ ),
+ n_sds_above_mean_thresh=cls._custom_widget(
+ "n_sds_above_mean_thresh", custom_label="Threshold"
+ ),
+ soma_spread_factor=cls._custom_widget(
+ "soma_spread_factor", custom_label="Cell spread"
+ ),
+ max_cluster_size=cls._custom_widget(
+ "max_cluster_size",
+ custom_label="Max cluster",
+ min=0,
+ max=10000000,
+ ),
+ )
+
+
+@dataclass
+class ClassificationInputs(InputContainer):
+ """Container for classification inputs."""
+
+ use_pre_trained_weights: bool = True
+ trained_model: Optional[Path] = Path.home()
+
+ def as_core_arguments(self) -> dict:
+ args = super().as_core_arguments()
+ del args["use_pre_trained_weights"]
+ return args
+
+ @classmethod
+ def widget_representation(cls) -> dict:
+ return dict(
+ classification_options=html_label_widget("Classification:"),
+ use_pre_trained_weights=dict(
+ value=cls.defaults()["use_pre_trained_weights"]
+ ),
+ trained_model=dict(value=cls.defaults()["trained_model"]),
+ )
+
+
+@dataclass
+class MiscInputs(InputContainer):
+ """Container for miscellaneous inputs."""
+
+ start_plane: int = 0
+ end_plane: int = 0
+ n_free_cpus: int = 2
+ analyse_local: bool = False
+ debug: bool = False
+
+ def as_core_arguments(self) -> dict:
+ misc_input_dict = super().as_core_arguments()
+ del misc_input_dict["debug"]
+ del misc_input_dict["analyse_local"]
+ return misc_input_dict
+
+ @classmethod
+ def widget_representation(cls) -> dict:
+ return dict(
+ misc_options=html_label_widget("Miscellaneous:"),
+ start_plane=cls._custom_widget("start_plane", min=0, max=100000),
+ end_plane=cls._custom_widget("end_plane", min=0, max=100000),
+ n_free_cpus=cls._custom_widget(
+ "n_free_cpus", custom_label="Number of free CPUs"
+ ),
+ analyse_local=dict(value=cls.defaults()["analyse_local"]),
+ debug=dict(value=cls.defaults()["debug"]),
+ )
diff --git a/cellfinder/napari/detect/thread_worker.py b/cellfinder/napari/detect/thread_worker.py
new file mode 100644
index 00000000..ea44dded
--- /dev/null
+++ b/cellfinder/napari/detect/thread_worker.py
@@ -0,0 +1,78 @@
+from napari.qt.threading import WorkerBase, WorkerBaseSignals
+from qtpy.QtCore import Signal
+
+from cellfinder.core.main import main as cellfinder_run
+
+from .detect_containers import (
+ ClassificationInputs,
+ DataInputs,
+ DetectionInputs,
+ MiscInputs,
+)
+
+
+class MyWorkerSignals(WorkerBaseSignals):
+ """
+ Signals used by the Worker class below.
+ """
+
+ # Emits (label, max, value) for the progress bar
+ update_progress_bar = Signal(str, int, int)
+
+
+class Worker(WorkerBase):
+ """
+ Runs cellfinder in a separate thread, to prevent GUI blocking.
+
+ Also handles callbacks between the worker thread and main napari GUI thread
+ to update a progress bar.
+ """
+
+ def __init__(
+ self,
+ data_inputs: DataInputs,
+ detection_inputs: DetectionInputs,
+ classification_inputs: ClassificationInputs,
+ misc_inputs: MiscInputs,
+ ):
+ super().__init__(SignalsClass=MyWorkerSignals)
+ self.data_inputs = data_inputs
+ self.detection_inputs = detection_inputs
+ self.classification_inputs = classification_inputs
+ self.misc_inputs = misc_inputs
+
+ def work(self) -> list:
+ self.update_progress_bar.emit("Setting up detection...", 1, 0)
+
+ def detect_callback(plane: int) -> None:
+ self.update_progress_bar.emit(
+ "Detecting cells",
+ self.data_inputs.nplanes,
+ plane + 1,
+ )
+
+ def detect_finished_callback(points: list) -> None:
+ self.npoints_detected = len(points)
+ self.update_progress_bar.emit("Setting up classification...", 1, 0)
+
+ def classify_callback(batch: int) -> None:
+ self.update_progress_bar.emit(
+ "Classifying cells",
+ # Default cellfinder-core batch size is 32. This seems to give
+ # a slight underestimate of the number of batches though, so
+ # allow for batch number to go over this
+ max(self.npoints_detected // 32 + 1, batch + 1),
+ batch + 1,
+ )
+
+ result = cellfinder_run(
+ **self.data_inputs.as_core_arguments(),
+ **self.detection_inputs.as_core_arguments(),
+ **self.classification_inputs.as_core_arguments(),
+ **self.misc_inputs.as_core_arguments(),
+ detect_callback=detect_callback,
+ classify_callback=classify_callback,
+ detect_finished_callback=detect_finished_callback,
+ )
+ self.update_progress_bar.emit("Finished classification", 1, 1)
+ return result
diff --git a/cellfinder/napari/images/brainglobe.png b/cellfinder/napari/images/brainglobe.png
new file mode 100644
index 00000000..427bdaba
Binary files /dev/null and b/cellfinder/napari/images/brainglobe.png differ
diff --git a/cellfinder/napari/input_container.py b/cellfinder/napari/input_container.py
new file mode 100644
index 00000000..5afa2937
--- /dev/null
+++ b/cellfinder/napari/input_container.py
@@ -0,0 +1,64 @@
+from abc import abstractmethod
+from dataclasses import asdict, dataclass
+from typing import Optional
+
+
+@dataclass
+class InputContainer:
+ """Base for classes that contain inputs
+
+ Intended to be derived to group specific related widget inputs (e.g from
+ the same widget section) into a container. Derived classes should be
+ Python data classes.
+
+ Enforces common interfaces for
+ - how to get default values for the inputs
+ - how inputs are passed to cellfinder core
+ - how the inputs are shown in the widget
+ """
+
+ @classmethod
+ def defaults(cls) -> dict:
+ """Returns default values of this class's fields as a dict."""
+ # Derived classes are not expected to be particularly
+ # slow to instantiate, so use the default constructor
+ # to avoid code repetition.
+ return asdict(cls())
+
+ @abstractmethod
+ def as_core_arguments(self) -> dict:
+ """Determines how dataclass fields are passed to cellfinder-core.
+
+ The implementation provided here can be re-used in derived classes, if
+ convenient.
+ """
+ # note that asdict returns a new instance of a dict,
+ # so any subsequent modifications of this dict won't affect the class
+ # instance
+ return asdict(self)
+
+ @classmethod
+ def _custom_widget(
+ cls, key: str, custom_label: Optional[str] = None, **kwargs
+ ) -> dict:
+ """
+ Represents a field, given by key, as a formatted widget with the
+ field's default value.
+
+ The widget label is the capitalized key by default, with underscores
+ replaced by spaces, unless custom_label is specified. Keyword
+ arguments like step, min, max, ... are passed to napari underneath.
+ """
+ label = (
+ key.replace("_", " ").capitalize()
+ if custom_label is None
+ else custom_label
+ )
+ value = cls.defaults()[key]
+ return dict(value=value, label=label, **kwargs)
+
+ @classmethod
+ @abstractmethod
+ def widget_representation(cls) -> dict:
+ """What the class will look like as a napari widget"""
+ pass
diff --git a/cellfinder/napari/napari.yaml b/cellfinder/napari/napari.yaml
new file mode 100644
index 00000000..903f090b
--- /dev/null
+++ b/cellfinder/napari/napari.yaml
@@ -0,0 +1,32 @@
+name: cellfinder
+schema_version: 0.1.0
+contributions:
+ commands:
+ - id: cellfinder.napari.detect_widget
+ title: Create Cell detection
+ python_name: cellfinder.napari.detect:detect_widget
+
+ - id: cellfinder.napari.training_widget
+ title: Create Train network
+ python_name: cellfinder.napari.train:training_widget
+
+ - id: cellfinder.napari.CurationWidget
+ title: Create Curation
+ python_name: cellfinder.napari.curation:CurationWidget
+
+ - id: cellfinder.napari.SampleData
+ title: Sample data
+ python_name: cellfinder.napari.sample_data:load_sample
+
+ widgets:
+ - command: cellfinder.napari.detect_widget
+ display_name: Cell detection
+ - command: cellfinder.napari.training_widget
+ display_name: Train network
+ - command: cellfinder.napari.CurationWidget
+ display_name: Curation
+
+ sample_data:
+ - key: sample
+ display_name: Sample data
+ command: cellfinder.napari.SampleData
diff --git a/cellfinder/napari/sample_data.py b/cellfinder/napari/sample_data.py
new file mode 100644
index 00000000..e42368e6
--- /dev/null
+++ b/cellfinder/napari/sample_data.py
@@ -0,0 +1,29 @@
+from typing import List
+
+import numpy as np
+import pooch
+from napari.types import LayerData
+from skimage.io import imread
+
+base_url = (
+ "https://raw.githubusercontent.com/brainglobe/cellfinder/"
+ "master/tests/data/integration/detection/crop_planes"
+)
+
+
+def load_sample() -> List[LayerData]:
+ """
+ Load some sample data.
+ """
+ layers = []
+ for ch, name in zip([1, 0], ["Background", "Signal"]):
+ data = []
+ for i in range(30):
+ url = f"{base_url}/ch{ch}/ch{ch}{str(i).zfill(4)}.tif"
+ file = pooch.retrieve(url=url, known_hash=None)
+ data.append(imread(file))
+
+ data = np.stack(data, axis=0)
+ layers.append((data, {"name": name}))
+
+ return layers
diff --git a/cellfinder/napari/train/__init__.py b/cellfinder/napari/train/__init__.py
new file mode 100644
index 00000000..48daa28a
--- /dev/null
+++ b/cellfinder/napari/train/__init__.py
@@ -0,0 +1 @@
+from .train import training_widget
diff --git a/cellfinder/napari/train/train.py b/cellfinder/napari/train/train.py
new file mode 100644
index 00000000..54d985a4
--- /dev/null
+++ b/cellfinder/napari/train/train.py
@@ -0,0 +1,178 @@
+from pathlib import Path
+from typing import Optional
+
+from magicgui import magicgui
+from magicgui.widgets import FunctionGui, PushButton
+from napari.qt.threading import thread_worker
+from napari.utils.notifications import show_info
+
+from cellfinder.core.train.train_yml import run as train_yml
+from cellfinder.napari.utils import (
+ header_label_widget,
+ html_label_widget,
+ widget_header,
+)
+
+from .train_containers import (
+ MiscTrainingInputs,
+ OptionalNetworkInputs,
+ OptionalTrainingInputs,
+ TrainingDataInputs,
+)
+
+
+@thread_worker
+def run_training(
+ training_data_inputs: TrainingDataInputs,
+ optional_network_inputs: OptionalNetworkInputs,
+ optional_training_inputs: OptionalTrainingInputs,
+ misc_training_inputs: MiscTrainingInputs,
+):
+ print("Running training")
+ train_yml(
+ **training_data_inputs.as_core_arguments(),
+ **optional_network_inputs.as_core_arguments(),
+ **optional_training_inputs.as_core_arguments(),
+ **misc_training_inputs.as_core_arguments(),
+ )
+ print("Finished!")
+
+
+def training_widget() -> FunctionGui:
+ @magicgui(
+ header=header_label_widget,
+ training_label=html_label_widget("Network training", tag="h3"),
+ **TrainingDataInputs.widget_representation(),
+ **OptionalNetworkInputs.widget_representation(),
+ **OptionalTrainingInputs.widget_representation(),
+ **MiscTrainingInputs.widget_representation(),
+ call_button=True,
+ reset_button=dict(widget_type="PushButton", text="Reset defaults"),
+ )
+ def widget(
+ header: dict,
+ training_label: dict,
+ data_options: dict,
+ yaml_files: Path,
+ output_directory: Path,
+ network_options: dict,
+ trained_model: Optional[Path],
+ model_weights: Optional[Path],
+ model_depth: str,
+ pretrained_model: str,
+ training_options: dict,
+ continue_training: bool,
+ augment: bool,
+ tensorboard: bool,
+ save_weights: bool,
+ save_checkpoints: bool,
+ save_progress: bool,
+ epochs: int,
+ learning_rate: float,
+ batch_size: int,
+ test_fraction: float,
+ misc_options: dict,
+ number_of_free_cpus: int,
+ reset_button: PushButton,
+ ):
+ """
+ Parameters
+ ----------
+ yaml_files : Path
+ YAML files containing paths to training data
+ output_directory : Path
+ Directory to save the output trained model
+ trained_model : Optional[Path]
+ Existing pre-trained model
+ model_weights : Optional[Path]
+ Existing pre-trained model weights
+ Should be set along with "Model depth"
+ model_depth : str
+ ResNet model depth (as per He et al. (2015))
+ pretrained_model : str
+ Which pre-trained model to use
+ (Supplied with cellfinder)
+ continue_training : bool
+ Continue training from an existing trained model
+ If no trained model or model weights are specified,
+ this will continue from the pretrained model
+ augment : bool
+ Augment the training data to improve generalisation
+ tensorboard : bool
+ Log to output_directory/tensorboard
+ save_weights : bool
+ Only store the model weights, and not the full model
+ Useful to save storage space
+ save_checkpoints : bool
+ Store the model at intermediate points during training
+ save_progress : bool
+ Save training progress to a .csv file
+ epochs : int
+ Number of training epochs
+ (How many times to use each training data point)
+ learning_rate : float
+ Learning rate for training the model
+ batch_size : int
+ Training batch size
+ test_fraction : float
+ Fraction of training data to use for validation
+ number_of_free_cpus : int
+ How many CPU cores to leave free
+ reset_button : PushButton
+ Reset parameters to default
+ """
+ trained_model = None if trained_model == Path.home() else trained_model
+ model_weights = None if model_weights == Path.home() else model_weights
+
+ training_data_inputs = TrainingDataInputs(yaml_files, output_directory)
+
+ optional_network_inputs = OptionalNetworkInputs(
+ trained_model,
+ model_weights,
+ model_depth,
+ pretrained_model,
+ )
+
+ optional_training_inputs = OptionalTrainingInputs(
+ continue_training,
+ augment,
+ tensorboard,
+ save_weights,
+ save_checkpoints,
+ save_progress,
+ epochs,
+ learning_rate,
+ batch_size,
+ test_fraction,
+ )
+
+ misc_training_inputs = MiscTrainingInputs(number_of_free_cpus)
+
+ if yaml_files[0] == Path.home(): # type: ignore
+ show_info("Please select a YAML file for training")
+ else:
+ worker = run_training(
+ training_data_inputs,
+ optional_network_inputs,
+ optional_training_inputs,
+ misc_training_inputs,
+ )
+ worker.start()
+
+ widget.header.value = widget_header
+ widget.header.native.setOpenExternalLinks(True)
+
+ @widget.reset_button.changed.connect
+ def restore_defaults():
+ defaults = {
+ **TrainingDataInputs.defaults(),
+ **OptionalNetworkInputs.defaults(),
+ **OptionalTrainingInputs.defaults(),
+ **MiscTrainingInputs.defaults(),
+ }
+ for name, value in defaults.items():
+ # ignore fields with no default
+ if value is not None:
+ getattr(widget, name).value = value
+
+ return widget
diff --git a/cellfinder/napari/train/train_containers.py b/cellfinder/napari/train/train_containers.py
new file mode 100644
index 00000000..73c6ae1e
--- /dev/null
+++ b/cellfinder/napari/train/train_containers.py
@@ -0,0 +1,127 @@
+from dataclasses import dataclass
+from pathlib import Path
+from typing import Optional
+
+from magicgui.types import FileDialogMode
+
+from cellfinder.core.download.models import model_weight_urls
+from cellfinder.core.train.train_yml import models
+from cellfinder.napari.input_container import InputContainer
+from cellfinder.napari.utils import html_label_widget
+
+
+@dataclass
+class TrainingDataInputs(InputContainer):
+ """Container for Training Data input widgets"""
+
+ yaml_files: Path = Path.home()
+ output_directory: Path = Path.home()
+
+ def as_core_arguments(self) -> dict:
+ arguments = super().as_core_arguments()
+ arguments["output_dir"] = arguments.pop("output_directory")
+ arguments["yaml_file"] = arguments.pop("yaml_files")
+ return arguments
+
+ @classmethod
+ def widget_representation(cls) -> dict:
+ return dict(
+ data_options=html_label_widget("Training Data:"),
+ yaml_files=cls._custom_widget(
+ "yaml_files",
+ custom_label="YAML files",
+ mode=FileDialogMode.EXISTING_FILES,
+ filter="*.yml",
+ ),
+ output_directory=cls._custom_widget(
+ "output_directory", mode=FileDialogMode.EXISTING_DIRECTORY
+ ),
+ )
+
+
+@dataclass
+class OptionalNetworkInputs(InputContainer):
+ """Container for Optional Network input widgets"""
+
+ trained_model: Optional[Path] = Path.home()
+ model_weights: Optional[Path] = Path.home()
+ model_depth: str = list(models.keys())[2]
+ pretrained_model: str = str(list(model_weight_urls.keys())[0])
+
+ def as_core_arguments(self) -> dict:
+ arguments = super().as_core_arguments()
+ arguments["model"] = arguments.pop("pretrained_model")
+ arguments["network_depth"] = arguments.pop("model_depth")
+ return arguments
+
+ @classmethod
+ def widget_representation(cls) -> dict:
+ return dict(
+ network_options=html_label_widget("Network (optional)"),
+ trained_model=cls._custom_widget("trained_model"),
+ model_weights=cls._custom_widget("model_weights"),
+ model_depth=cls._custom_widget(
+ "model_depth", choices=list(models.keys())
+ ),
+ pretrained_model=cls._custom_widget(
+ "pretrained_model",
+ choices=list(model_weight_urls.keys()),
+ ),
+ )
+
+
+@dataclass
+class OptionalTrainingInputs(InputContainer):
+ continue_training: bool = False
+ augment: bool = True
+ tensorboard: bool = False
+ save_weights: bool = False
+ save_checkpoints: bool = True
+ save_progress: bool = True
+ epochs: int = 100
+ learning_rate: float = 1e-4
+ batch_size: int = 16
+ test_fraction: float = 0.1
+
+ def as_core_arguments(self) -> dict:
+ arguments = super().as_core_arguments()
+ arguments["no_augment"] = not arguments.pop("augment")
+ arguments["no_save_checkpoints"] = not arguments.pop(
+ "save_checkpoints"
+ )
+ return arguments
+
+ @classmethod
+ def widget_representation(cls) -> dict:
+ return dict(
+ training_options=html_label_widget("Training (optional)"),
+ continue_training=cls._custom_widget("continue_training"),
+ augment=cls._custom_widget("augment"),
+ tensorboard=cls._custom_widget("tensorboard"),
+ save_weights=cls._custom_widget("save_weights"),
+ save_checkpoints=cls._custom_widget("save_checkpoints"),
+ save_progress=cls._custom_widget("save_progress"),
+ epochs=cls._custom_widget("epochs"),
+ learning_rate=cls._custom_widget("learning_rate", step=1e-4),
+ batch_size=cls._custom_widget("batch_size"),
+ test_fraction=cls._custom_widget(
+ "test_fraction", step=0.05, min=0.05, max=0.95
+ ),
+ )
+
+
+@dataclass
+class MiscTrainingInputs(InputContainer):
+ number_of_free_cpus: int = 2
+
+ def as_core_arguments(self) -> dict:
+ return dict(n_free_cpus=self.number_of_free_cpus)
+
+ @classmethod
+ def widget_representation(cls) -> dict:
+ return dict(
+ misc_options=html_label_widget("Misc (optional):"),
+ number_of_free_cpus=cls._custom_widget(
+ "number_of_free_cpus", custom_label="Number of free CPUs"
+ ),
+ )
diff --git a/cellfinder/napari/utils.py b/cellfinder/napari/utils.py
new file mode 100644
index 00000000..6c82a4b2
--- /dev/null
+++ b/cellfinder/napari/utils.py
@@ -0,0 +1,180 @@
+from typing import Callable, List, Optional, Tuple
+
+import napari
+import numpy as np
+import pandas as pd
+from brainglobe_utils.cells.cells import Cell
+from pkg_resources import resource_filename
+from qtpy.QtWidgets import (
+ QComboBox,
+ QLabel,
+ QLayout,
+ QMessageBox,
+ QPushButton,
+ QWidget,
+)
+
+brainglobe_logo = resource_filename(
+ "cellfinder", "napari/images/brainglobe.png"
+)
+
+
+widget_header = """
+Efficient cell detection in large images.
+Website
+Documentation
+Source
+Citation
+For help, hover the cursor over each parameter.
+""" # noqa: E501
+
+
+def html_label_widget(label: str, *, tag: str = "b") -> dict:
+ """
+ Create a HMTL label for use with magicgui.
+ """
+ return dict(
+ widget_type="Label",
+ label=f"<{tag}>{label}",
+ )
+
+
+header_label_widget = html_label_widget(
+ f"""
+ +
+
cellfinder
+""",
+ tag="h1",
+)
+
+
+def add_layers(points: List[Cell], viewer: napari.Viewer) -> None:
+ """
+ Adds classified cell candidates as two separate point layers to the napari
+ viewer.
+ """
+ detected, rejected = cells_to_array(points)
+
+ viewer.add_points(
+ rejected,
+ name="Rejected",
+ size=15,
+ n_dimensional=True,
+ opacity=0.6,
+ symbol="ring",
+ face_color="lightskyblue",
+ visible=False,
+ metadata=dict(point_type=Cell.UNKNOWN),
+ )
+ viewer.add_points(
+ detected,
+ name="Detected",
+ size=15,
+ n_dimensional=True,
+ opacity=0.6,
+ symbol="ring",
+ face_color="lightgoldenrodyellow",
+ metadata=dict(point_type=Cell.CELL),
+ )
+
+
+def cells_df_as_np(
+ cells_df: pd.DataFrame,
+ new_order: List[int] = [2, 1, 0],
+ type_column: str = "type",
+) -> np.ndarray:
+ """
+ Convert a dataframe to an array, dropping *type_column* and re-ordering
+ the columns with *new_order*.
+ """
+ cells_df = cells_df.drop(columns=[type_column])
+ cells = cells_df[cells_df.columns[new_order]]
+ cells = cells.to_numpy()
+ return cells
+
+
+def cells_to_array(cells: List[Cell]) -> Tuple[np.ndarray, np.ndarray]:
+ df = pd.DataFrame([c.to_dict() for c in cells])
+ points = cells_df_as_np(df[df["type"] == Cell.CELL])
+ rejected = cells_df_as_np(df[df["type"] == Cell.UNKNOWN])
+ return points, rejected
+
+
+def add_combobox(
+ layout: QLayout,
+ label: str,
+ items: List[str],
+ row: int,
+ column: int = 0,
+ label_stack: bool = False,
+ callback=None,
+ width: int = 150,
+) -> Tuple[QComboBox, Optional[QLabel]]:
+ """
+ Add a selection box to *layout*.
+ """
+ if label_stack:
+ combobox_row = row + 1
+ combobox_column = column
+ else:
+ combobox_row = row
+ combobox_column = column + 1
+ combobox = QComboBox()
+ combobox.addItems(items)
+ if callback:
+ combobox.currentIndexChanged.connect(callback)
+ combobox.setMaximumWidth = width
+
+ if label is not None:
+ combobox_label = QLabel(label)
+ combobox_label.setMaximumWidth = width
+ layout.addWidget(combobox_label, row, column)
+ else:
+ combobox_label = None
+
+ layout.addWidget(combobox, combobox_row, combobox_column)
+ return combobox, combobox_label
+
+
+def add_button(
+ label: str,
+ layout: QLayout,
+ connected_function: Callable,
+ row: int,
+ column: int = 0,
+ visibility: bool = True,
+ minimum_width: int = 0,
+ alignment: str = "center",
+) -> QPushButton:
+ """
+ Add a button to *layout*.
+ """
+ button = QPushButton(label)
+ if alignment == "center":
+ pass
+ elif alignment == "left":
+ button.setStyleSheet("QPushButton { text-align: left; }")
+ elif alignment == "right":
+ button.setStyleSheet("QPushButton { text-align: right; }")
+
+ button.setVisible(visibility)
+ button.setMinimumWidth(minimum_width)
+ layout.addWidget(button, row, column)
+ button.clicked.connect(connected_function)
+ return button
+
+
+def display_question(widget: QWidget, title: str, message: str) -> bool:
+ """
+ Display a warning in a pop up that informs about overwriting files.
+ """
+ message_reply = QMessageBox.question(
+ widget,
+ title,
+ message,
+ QMessageBox.Yes | QMessageBox.Cancel,
+ )
+ if message_reply == QMessageBox.Yes:
+ return True
+ else:
+ return False
diff --git a/examples/show_detection_sample.py b/examples/show_detection_sample.py
new file mode 100644
index 00000000..3a44f7d1
--- /dev/null
+++ b/examples/show_detection_sample.py
@@ -0,0 +1,27 @@
+"""
+Load and show sample data
+=========================
+This example:
+- loads some sample data
+- adds the data to a napari viewer
+- loads the cellfinder-napari cell detection plugin
+- opens the napari viewer
+"""
+import napari
+
+from cellfinder.napari.sample_data import load_sample
+
+viewer = napari.Viewer()
+# Open plugin
+viewer.window.add_plugin_dock_widget(
+ plugin_name="cellfinder", widget_name="Cell detection"
+)
+# Add sample data layers
+for layer in load_sample():
+ viewer.add_layer(napari.layers.Image(layer[0], **layer[1]))
+
+
+if __name__ == "__main__":
+ # The napari event loop needs to be run under here to allow the window
+ # to be spawned from a Python script
+ napari.run()
diff --git a/pyproject.toml b/pyproject.toml
index 85bbb955..c77fde52 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,16 +1,19 @@
[project]
-name = "cellfinder-core"
+name = "cellfinder"
description = "Automated 3D cell detection in large microscopy images"
readme = "README.md"
+license = { text = "BSD-3-Clause" }
authors = [
- {name = "Adam Tyson, Christian Niedworok, Charly Rousseau", email = "code@adamltyson.com"},
+ { name = "Adam Tyson, Christian Niedworok, Charly Rousseau", email = "code@adamltyson.com" },
]
classifiers = [
- "Development Status :: 3 - Alpha",
+ "Development Status :: 4 - Beta",
+ "Framework :: napari",
"Intended Audience :: Developers",
"Intended Audience :: Science/Research",
"Operating System :: OS Independent",
"Programming Language :: Python",
+ "Programming Language :: Python :: 3",
"Programming Language :: Python :: 3.8",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
@@ -18,9 +21,9 @@ classifiers = [
]
requires-python = ">=3.8"
dependencies = [
+ "brainglobe-utils",
"dask[array]",
"fancylog>=0.0.7",
- "brainglobe-utils",
"natsort",
"numba",
"numpy",
@@ -32,106 +35,76 @@ dependencies = [
"tifffile",
"tqdm",
]
-dynamic = ['version']
+dynamic = ["version"]
-[project.urls]
-Homepage = "https://brainglobe.info"
-"Source Code" = "https://github.com/brainglobe/cellfinder-core"
-"Bug Tracker" = "https://github.com/brainglobe/cellfinder-core/issues"
-Documentation = "https://brainglobe.info/documentation/cellfinder/index.html"
+[project.entry-points."napari.manifest"]
+cellfinder = "cellfinder.napari:napari.yaml"
[project.optional-dependencies]
dev = [
"black",
- "gitpython",
"pre-commit",
"pyinstrument",
- "pytest",
"pytest-cov",
"pytest-lazy-fixture",
"pytest-mock",
+ "pytest-qt",
"pytest-timeout",
+ "pytest",
"tox",
]
+napari = [
+ "brainglobe-napari-io",
+ "magicgui",
+ "napari-ndtiffs",
+ "napari-plugin-engine >= 0.1.4",
+ "napari[all]",
+ "pooch >= 1",
+ "qtpy",
+]
[project.scripts]
-cellfinder_download = "cellfinder_core.download.cli:main"
-cellfinder_train = "cellfinder_core.train.train_yml:cli"
+cellfinder_download = "cellfinder.core.download.cli:main"
+cellfinder_train = "cellfinder.core.train.train_yml:cli"
+
+[project.urls]
+Homepage = "https://brainglobe.info/documentation/cellfinder/index.html"
+"Source Code" = "https://github.com/brainglobe/cellfinder-core"
+"Bug Tracker" = "https://github.com/brainglobe/cellfinder-core/issues"
+Documentation = "https://brainglobe.info/documentation/cellfinder/index.html"
+"User Support" = "https://forum.image.sc/tag/brainglobe"
[build-system]
-requires = [
- "setuptools>=45",
- "wheel",
- "setuptools_scm[toml]>=6.2",
-]
+requires = ["setuptools>=45", "wheel", "setuptools_scm[toml]>=6.2"]
build-backend = 'setuptools.build_meta'
-[tool.setuptools]
-include-package-data = true
-
-[tool.setuptools_scm]
-
-[tool.cibuildwheel]
-build = "cp38-* cp39-* cp310-*"
-
-[tool.cibuildwheel.macos]
-archs = ["x86_64", "arm64"]
-
-[tool.pytest.ini_options]
-addopts = "--cov=cellfinder_core --cov-report=xml"
-filterwarnings = [
- "error",
- # Raised by tensorflow; should be removed when tensorflow 2.12.0 is released. Fix is:
- # https://github.com/tensorflow/tensorflow/commit/b23c5750c9f35a87872793eef7c56e74ec55d4a7
- "ignore:`np.bool8` is a deprecated alias for `np.bool_`",
- # See https://github.com/numba/numba/issues/8676
- "ignore:.*:numba.core.errors.NumbaTypeSafetyWarning"
-]
-markers = [
- "slow: marks tests as slow (deselect with '-m \"not slow\"')",
- "serial",
-]
-log_level = "WARNING"
-
[tool.black]
target-version = ['py38', 'py39', 'py310']
skip-string-normalization = false
line-length = 79
+[tool.cibuildwheel.macos]
+archs = ["x86_64", "arm64"]
+
[tool.ruff]
line-length = 79
-exclude = ["__init__.py","build",".eggs"]
+exclude = ["__init__.py", "build", ".eggs"]
select = ["I", "E", "F"]
fix = true
[tool.ruff.isort]
-known-first-party = ["cellfinder_core"]
+known-first-party = ["cellfinder"]
-[tool.mypy]
+[tool.setuptools]
+include-package-data = true
-[[tool.mypy.overrides]]
-module = [
- "fancylog.*",
- "brainglobe_utils.*",
- "natsort.*",
- "numba.*",
- "tensorflow.*",
- "tifffile.*",
- "pyinstrument.*",
- "pytest.*",
- "pytest_lazyfixture.*",
- "scipy.*",
- "skimage.*",
- "sklearn.*",
- "cellfinder_core.tools.prep.*",
-]
-ignore_missing_imports = true
+[tool.setuptools.packages.find]
+include = ["cellfinder*"]
-[[tool.mypy.overrides]]
-module = [
- "cellfinder_core.detect.*",
- "cellfinder_core.classify.*",
-]
-disallow_untyped_defs = true
-disallow_incomplete_defs = true
-disallow_untyped_calls = true
+[tool.setuptools.package-data]
+include = ["cellfinder*"]
+
+[tool.setuptools_scm]
+
+[tool.cibuildwheel]
+build = "cp38-* cp39-* cp310-*"
diff --git a/resources/cellfinder-napari.gif b/resources/cellfinder-napari.gif
new file mode 100644
index 00000000..c3136fba
Binary files /dev/null and b/resources/cellfinder-napari.gif differ
diff --git a/tests/tests/__init__.py b/tests/core/__init__.py
similarity index 100%
rename from tests/tests/__init__.py
rename to tests/core/__init__.py
diff --git a/tests/tests/conftest.py b/tests/core/conftest.py
similarity index 94%
rename from tests/tests/conftest.py
rename to tests/core/conftest.py
index a65a809f..f05ec88a 100644
--- a/tests/tests/conftest.py
+++ b/tests/core/conftest.py
@@ -5,8 +5,8 @@
import pytest
from skimage.filters import gaussian
-from cellfinder_core.download import models
-from cellfinder_core.tools.prep import DEFAULT_INSTALL_PATH
+from cellfinder.core.download import models
+from cellfinder.core.tools.prep import DEFAULT_INSTALL_PATH
@pytest.fixture(scope="session")
diff --git a/tests/tests/test_integration/__init__.py b/tests/core/test_integration/__init__.py
similarity index 100%
rename from tests/tests/test_integration/__init__.py
rename to tests/core/test_integration/__init__.py
diff --git a/tests/tests/test_integration/test_detection.py b/tests/core/test_integration/test_detection.py
similarity index 96%
rename from tests/tests/test_integration/test_detection.py
rename to tests/core/test_integration/test_detection.py
index f16a437b..ed289d10 100644
--- a/tests/tests/test_integration/test_detection.py
+++ b/tests/core/test_integration/test_detection.py
@@ -6,8 +6,8 @@
import pytest
from brainglobe_utils.general.system import get_num_processes
-from cellfinder_core.main import main
-from cellfinder_core.tools.IO import read_with_dask
+from cellfinder.core.main import main
+from cellfinder.core.tools.IO import read_with_dask
data_dir = os.path.join(
os.getcwd(), "tests", "data", "integration", "detection"
@@ -86,7 +86,7 @@ def test_detection_small_planes(
n_planes = 2
# Don't want to bother classifying in this test, so mock classifcation
- mocker.patch("cellfinder_core.classify.classify.main")
+ mocker.patch("cellfinder.core.classify.classify.main")
pytest.mark.skipif(
nproc < n_planes,
diff --git a/tests/tests/test_integration/test_detection_structure_splitting.py b/tests/core/test_integration/test_detection_structure_splitting.py
similarity index 92%
rename from tests/tests/test_integration/test_detection_structure_splitting.py
rename to tests/core/test_integration/test_detection_structure_splitting.py
index 518d2a9e..87ae7cb2 100644
--- a/tests/tests/test_integration/test_detection_structure_splitting.py
+++ b/tests/core/test_integration/test_detection_structure_splitting.py
@@ -10,8 +10,8 @@
import pytest
-from cellfinder_core.main import main
-from cellfinder_core.tools.IO import read_with_dask
+from cellfinder.core.main import main
+from cellfinder.core.tools.IO import read_with_dask
data_dir = os.path.join(
os.getcwd(), "tests", "data", "integration", "detection"
diff --git a/tests/tests/test_integration/test_train.py b/tests/core/test_integration/test_train.py
similarity index 92%
rename from tests/tests/test_integration/test_train.py
rename to tests/core/test_integration/test_train.py
index d611d222..0d4f5c38 100644
--- a/tests/tests/test_integration/test_train.py
+++ b/tests/core/test_integration/test_train.py
@@ -3,7 +3,7 @@
import pytest
-from cellfinder_core.train.train_yml import cli as train_run
+from cellfinder.core.train.train_yml import cli as train_run
data_dir = os.path.join(
os.getcwd(), "tests", "data", "integration", "training"
diff --git a/tests/tests/test_unit/__init__.py b/tests/core/test_unit/__init__.py
similarity index 100%
rename from tests/tests/test_unit/__init__.py
rename to tests/core/test_unit/__init__.py
diff --git a/tests/tests/test_unit/test_detect/__init__.py b/tests/core/test_unit/test_detect/__init__.py
similarity index 100%
rename from tests/tests/test_unit/test_detect/__init__.py
rename to tests/core/test_unit/test_detect/__init__.py
diff --git a/tests/tests/test_unit/test_detect/test_detect.py b/tests/core/test_unit/test_detect/test_detect.py
similarity index 90%
rename from tests/tests/test_unit/test_detect/test_detect.py
rename to tests/core/test_unit/test_detect/test_detect.py
index 149de53a..b74d2ed9 100644
--- a/tests/tests/test_unit/test_detect/test_detect.py
+++ b/tests/core/test_unit/test_detect/test_detect.py
@@ -1,6 +1,6 @@
import multiprocessing
-from cellfinder_core.detect.detect import _map_with_locks
+from cellfinder.core.detect.detect import _map_with_locks
def add_one(a: int) -> int:
diff --git a/tests/tests/test_unit/test_detect/test_filters/test_volume_filters/__init__.py b/tests/core/test_unit/test_detect/test_filters/test_volume_filters/__init__.py
similarity index 100%
rename from tests/tests/test_unit/test_detect/test_filters/test_volume_filters/__init__.py
rename to tests/core/test_unit/test_detect/test_filters/test_volume_filters/__init__.py
diff --git a/tests/tests/test_unit/test_detect/test_filters/test_volume_filters/test_structure_detection.py b/tests/core/test_unit/test_detect/test_filters/test_volume_filters/test_structure_detection.py
similarity index 98%
rename from tests/tests/test_unit/test_detect/test_filters/test_volume_filters/test_structure_detection.py
rename to tests/core/test_unit/test_detect/test_filters/test_volume_filters/test_structure_detection.py
index 8235874b..9895e2c9 100644
--- a/tests/tests/test_unit/test_detect/test_filters/test_volume_filters/test_structure_detection.py
+++ b/tests/core/test_unit/test_detect/test_filters/test_volume_filters/test_structure_detection.py
@@ -1,7 +1,7 @@
import numpy as np
import pytest
-from cellfinder_core.detect.filters.volume.structure_detection import (
+from cellfinder.core.detect.filters.volume.structure_detection import (
CellDetector,
Point,
get_non_zero_dtype_min,
diff --git a/tests/tests/test_unit/test_tools/__init__.py b/tests/core/test_unit/test_tools/__init__.py
similarity index 100%
rename from tests/tests/test_unit/test_tools/__init__.py
rename to tests/core/test_unit/test_tools/__init__.py
diff --git a/tests/tests/test_unit/test_tools/test_IO.py b/tests/core/test_unit/test_tools/test_IO.py
similarity index 91%
rename from tests/tests/test_unit/test_tools/test_IO.py
rename to tests/core/test_unit/test_tools/test_IO.py
index a649a577..523a69c6 100644
--- a/tests/tests/test_unit/test_tools/test_IO.py
+++ b/tests/core/test_unit/test_tools/test_IO.py
@@ -1,6 +1,6 @@
import dask.array as d_array
-from cellfinder_core.tools import IO
+from cellfinder.core.tools import IO
BRAIN_DIR = "tests/data/brain"
BRAIN_PATHS = f"{BRAIN_DIR}/brain_paths.txt"
diff --git a/tests/tests/test_unit/test_tools/test_geometry.py b/tests/core/test_unit/test_tools/test_geometry.py
similarity index 82%
rename from tests/tests/test_unit/test_tools/test_geometry.py
rename to tests/core/test_unit/test_tools/test_geometry.py
index 3d196f3a..9872d006 100644
--- a/tests/tests/test_unit/test_tools/test_geometry.py
+++ b/tests/core/test_unit/test_tools/test_geometry.py
@@ -1,6 +1,6 @@
import numpy as np
-import cellfinder_core.tools.geometry as geometry
+import cellfinder.core.tools.geometry as geometry
def test_make_sphere():
diff --git a/tests/tests/test_unit/test_tools/test_image_processing.py b/tests/core/test_unit/test_tools/test_image_processing.py
similarity index 94%
rename from tests/tests/test_unit/test_tools/test_image_processing.py
rename to tests/core/test_unit/test_tools/test_image_processing.py
index 62db4a67..64ad4891 100644
--- a/tests/tests/test_unit/test_tools/test_image_processing.py
+++ b/tests/core/test_unit/test_tools/test_image_processing.py
@@ -2,7 +2,7 @@
import numpy as np
-from cellfinder_core.tools import image_processing as img_tools
+from cellfinder.core.tools import image_processing as img_tools
def test_crop_center_2d():
diff --git a/tests/tests/test_unit/test_tools/test_system.py b/tests/core/test_unit/test_tools/test_system.py
similarity index 98%
rename from tests/tests/test_unit/test_tools/test_system.py
rename to tests/core/test_unit/test_tools/test_system.py
index 567f6555..40f87755 100644
--- a/tests/tests/test_unit/test_tools/test_system.py
+++ b/tests/core/test_unit/test_tools/test_system.py
@@ -6,7 +6,7 @@
from brainglobe_utils.general.exceptions import CommandLineInputError
from brainglobe_utils.general.system import ensure_directory_exists
-import cellfinder_core.tools.system as system
+import cellfinder.core.tools.system as system
data_dir = Path("tests", "data")
background_im_dir = os.path.join(data_dir, "background")
diff --git a/tests/tests/test_unit/test_tools/test_tools_general.py b/tests/core/test_unit/test_tools/test_tools_general.py
similarity index 97%
rename from tests/tests/test_unit/test_tools/test_tools_general.py
rename to tests/core/test_unit/test_tools/test_tools_general.py
index 7bac4e65..d3609cb4 100644
--- a/tests/tests/test_unit/test_tools/test_tools_general.py
+++ b/tests/core/test_unit/test_tools/test_tools_general.py
@@ -3,7 +3,7 @@
import numpy as np
import pytest
-import cellfinder_core.tools.tools as tools
+import cellfinder.core.tools.tools as tools
a = [1, "a", 10, 30]
b = [30, 10, "c", "d"]
diff --git a/tests/napari/__init__.py b/tests/napari/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/tests/napari/test_curation.py b/tests/napari/test_curation.py
new file mode 100644
index 00000000..f52141cc
--- /dev/null
+++ b/tests/napari/test_curation.py
@@ -0,0 +1,190 @@
+from pathlib import Path
+from unittest.mock import patch
+
+import napari
+import numpy as np
+import pytest
+from napari.layers import Image, Points
+
+from cellfinder.napari import sample_data
+from cellfinder.napari.curation import CurationWidget
+from cellfinder.napari.sample_data import load_sample
+
+
+@pytest.fixture
+def curation_widget(make_napari_viewer):
+ """
+ Create a viewer, add the curation widget, and return the widget.
+ The viewer can be accessed using ``widget.viewer``.
+ """
+ viewer = make_napari_viewer()
+ _, widget = viewer.window.add_plugin_dock_widget(
+ plugin_name="cellfinder", widget_name="Curation"
+ )
+ return widget
+
+
+def test_add_new_training_layers(curation_widget):
+ viewer = curation_widget.viewer
+ layers = viewer.layers
+ # Check that layers list starts off empty
+ assert len(layers) == 0
+ curation_widget.add_training_data()
+ assert len(layers) == 2
+
+ assert all(isinstance(layer, Points) for layer in layers)
+
+ assert layers[0].name == "Training data (cells)"
+ assert layers[1].name == "Training data (non cells)"
+
+
+def test_cell_marking(curation_widget, tmp_path):
+ """
+ Check that marking cells and non-cells works as expected.
+ """
+ widget = curation_widget
+ widget.add_training_data()
+ viewer = widget.viewer
+
+ cell_layer = widget.training_data_cell_layer
+ non_cell_layer = widget.training_data_non_cell_layer
+
+ # Check that no cells have been marked yet
+ assert all(
+ layer.data.shape == (0, 3) for layer in [cell_layer, non_cell_layer]
+ )
+
+ # Add a points layer to select points from
+ points = Points(
+ np.array([[16, 17, 18], [13, 14, 15]]), name="selection_points"
+ )
+ # Adding the layer automatically selects it in the layer list
+ viewer.add_layer(points)
+
+ # Select the first point, and add as a cell
+ points.selected_data = [0]
+ curation_widget.mark_as_cell()
+ assert np.array_equal(cell_layer.data, np.array([[16, 17, 18]]))
+ assert non_cell_layer.data.shape[0] == 0
+
+ # Select the second point, and add as a non-cell
+ points.selected_data = [1]
+ curation_widget.mark_as_non_cell()
+ assert np.array_equal(cell_layer.data, np.array([[16, 17, 18]]))
+ assert np.array_equal(non_cell_layer.data, np.array([[13, 14, 15]]))
+
+ # Add signal/background images to the viewer and widget
+ layer_data = sample_data.load_sample()
+ signal = Image(layer_data[0][0], **layer_data[0][1])
+ background = Image(layer_data[1][0], **layer_data[1][1])
+ widget.signal_layer = signal
+ widget.background_layer = background
+
+ widget.output_directory = tmp_path
+ widget.save_training_data(prompt_for_directory=False, block=True)
+
+ assert (tmp_path / "training.yml").exists()
+ # Check that two .tif files are saved for both cells and non_cells
+ assert len(list((tmp_path / "non_cells").glob("*.tif"))) == 2
+ assert len(list((tmp_path / "cells").glob("*.tif"))) == 2
+
+
+@pytest.fixture
+def valid_curation_widget(make_napari_viewer) -> CurationWidget:
+ """
+ Setup up a valid curation widget,
+ complete with training data layers and points,
+ and signal+background images.
+ """
+ viewer = make_napari_viewer()
+ image_layers = load_sample()
+ for layer in image_layers:
+ viewer.add_layer(napari.layers.Image(layer[0], **layer[1]))
+
+ num_dw = len(viewer.window._dock_widgets)
+ _, curation_widget = viewer.window.add_plugin_dock_widget(
+ plugin_name="cellfinder", widget_name="Curation"
+ )
+ assert len(viewer.window._dock_widgets) == num_dw + 1
+
+ curation_widget.add_training_data()
+ # Add a points layer to select points from
+ points = Points(
+ np.array([[16, 17, 18], [13, 14, 15]]), name="selection_points"
+ )
+ # Adding the layer automatically selects it in the layer list
+ viewer.add_layer(points)
+
+ # Select the first point, and add as a cell
+ points.selected_data = [0]
+ curation_widget.mark_as_cell()
+
+ # Select the second point, and add as a non-cell
+ points.selected_data = [1]
+ curation_widget.mark_as_non_cell()
+
+ curation_widget.signal_image_choice.setCurrentText("Signal")
+ curation_widget.background_image_choice.setCurrentText("Background")
+ curation_widget.set_signal_image()
+ curation_widget.set_background_image()
+ return curation_widget
+
+
+def test_check_image_data_for_extraction(valid_curation_widget):
+ """
+ Check valid curation widget has extractable data.
+ """
+ assert valid_curation_widget.check_image_data_for_extraction()
+
+
+def test_check_image_data_wrong_shape(valid_curation_widget):
+ """
+ Check curation widget shows expected user message if images don't have
+ identical shape.
+ """
+ with patch("cellfinder.napari.curation.show_info") as show_info:
+ signal_layer_with_wrong_shape = napari.layers.Image(
+ np.zeros(shape=(1, 1)), name="Wrong shape"
+ )
+ valid_curation_widget.viewer.add_layer(signal_layer_with_wrong_shape)
+ valid_curation_widget.signal_image_choice.setCurrentText("Wrong shape")
+ valid_curation_widget.set_signal_image()
+ valid_curation_widget.check_image_data_for_extraction()
+ show_info.assert_called_once_with(
+ "Please ensure both signal and background images are the "
+ "same size and shape."
+ )
+
+
+def test_check_image_data_missing_signal(valid_curation_widget):
+ """
+ Check curation widget shows expected user message if signal image is
+ missing.
+ """
+ with patch("cellfinder.napari.curation.show_info") as show_info:
+ valid_curation_widget.signal_layer = None
+ valid_curation_widget.check_image_data_for_extraction()
+ show_info.assert_called_once_with(
+ "Please ensure both signal and background images are loaded "
+ "into napari, and selected in the sidebar. "
+ )
+
+
+def test_is_data_extractable(curation_widget, valid_curation_widget):
+ """Check is_data_extractable works as expected."""
+ assert not curation_widget.is_data_extractable()
+ assert valid_curation_widget.is_data_extractable()
+
+
+def test_get_output_directory(valid_curation_widget):
+ """Check get_output_directory returns expected value."""
+ with patch(
+ "cellfinder.napari.curation.QFileDialog.getExistingDirectory"
+ ) as get_directory:
+ get_directory.return_value = ""
+ valid_curation_widget.get_output_directory()
+ assert valid_curation_widget.output_directory is None
+
+ get_directory.return_value = Path.home()
+ valid_curation_widget.get_output_directory()
+ assert valid_curation_widget.output_directory == Path.home()
diff --git a/tests/napari/test_detection.py b/tests/napari/test_detection.py
new file mode 100644
index 00000000..ea7c6fb5
--- /dev/null
+++ b/tests/napari/test_detection.py
@@ -0,0 +1,72 @@
+from unittest.mock import patch
+
+import napari
+import pytest
+
+from cellfinder.napari.detect import detect_widget
+from cellfinder.napari.detect.detect_containers import (
+ ClassificationInputs,
+ DataInputs,
+ DetectionInputs,
+ MiscInputs,
+)
+from cellfinder.napari.detect.thread_worker import Worker
+from cellfinder.napari.sample_data import load_sample
+
+
+@pytest.fixture
+def get_detect_widget(make_napari_viewer):
+ viewer = make_napari_viewer()
+ widget = detect_widget()
+ for layer in load_sample():
+ viewer.add_layer(napari.layers.Image(layer[0], **layer[1]))
+ _, widget = viewer.window.add_plugin_dock_widget(
+ plugin_name="cellfinder", widget_name="Cell detection"
+ )
+ return widget
+
+
+def test_detect_worker():
+ """
+ Smoke test to check that the detection worker runs
+ """
+ data = load_sample()
+ signal = data[0][0]
+ background = data[1][0]
+
+ worker = Worker(
+ DataInputs(signal_array=signal, background_array=background),
+ DetectionInputs(),
+ ClassificationInputs(trained_model=None),
+ MiscInputs(start_plane=0, end_plane=1),
+ )
+ worker.work()
+
+
+@pytest.mark.parametrize(
+ argnames="analyse_local",
+ argvalues=[True, False], # increase test coverage by covering both cases
+)
+def test_run_detect(get_detect_widget, analyse_local):
+ """
+ Test backend is called
+ """
+ with patch("cellfinder.napari.detect.detect.Worker") as worker:
+ get_detect_widget.analyse_local.value = analyse_local
+ get_detect_widget.call_button.clicked()
+ assert worker.called
+
+
+def test_run_detect_without_inputs():
+ """ """
+ with patch("cellfinder.napari.detect.detect.show_info") as show_info:
+ widget = (
+ detect_widget()
+ ) # won't have image layers, so should notice and show info
+ widget.call_button.clicked()
+ assert show_info.called
+
+
+def test_reset_defaults(get_detect_widget):
+ """Smoke test that restore defaults doesn't error."""
+ get_detect_widget.reset_button.clicked()
diff --git a/tests/napari/test_train_containers.py b/tests/napari/test_train_containers.py
new file mode 100644
index 00000000..57efaad8
--- /dev/null
+++ b/tests/napari/test_train_containers.py
@@ -0,0 +1,32 @@
+from inspect import signature
+
+import pytest
+
+from cellfinder.core.train.train_yml import run
+from cellfinder.napari.train.train_containers import (
+ MiscTrainingInputs,
+ OptionalNetworkInputs,
+ OptionalTrainingInputs,
+ TrainingDataInputs,
+)
+
+
+@pytest.mark.parametrize(
+ argnames="input_container",
+ argvalues=[
+ MiscTrainingInputs(),
+ OptionalNetworkInputs(),
+ OptionalTrainingInputs(),
+ TrainingDataInputs(),
+ ],
+)
+def test_core_args_passed(input_container):
+ """
+ Check that any keyword argument that napari passes
+ to the training backend actually is also expected by the backend
+ """
+ backend_signature = signature(run)
+ expected_kwargs_set = set(backend_signature.parameters.keys())
+ actual_kwargs_set = set(input_container.as_core_arguments().keys())
+ # check all actual keywords are in expected (but not the other way around.)
+ assert actual_kwargs_set <= expected_kwargs_set
diff --git a/tests/napari/test_training.py b/tests/napari/test_training.py
new file mode 100644
index 00000000..48ce1139
--- /dev/null
+++ b/tests/napari/test_training.py
@@ -0,0 +1,91 @@
+from pathlib import Path
+from unittest.mock import patch
+
+import pytest
+
+from cellfinder.napari.train.train import training_widget
+from cellfinder.napari.train.train_containers import (
+ MiscTrainingInputs,
+ OptionalNetworkInputs,
+ OptionalTrainingInputs,
+ TrainingDataInputs,
+)
+
+
+@pytest.fixture
+def get_training_widget(make_napari_viewer):
+ viewer = make_napari_viewer()
+ widget = training_widget()
+ _, widget = viewer.window.add_plugin_dock_widget(
+ plugin_name="cellfinder", widget_name="Train network"
+ )
+ viewer.window.add_dock_widget(widget)
+ return widget
+
+
+def test_reset_to_defaults(get_training_widget):
+ """
+ A simple test for the reset button.
+ Checks widgets of a few different types are reset as expected.
+ """
+ # change a few widgets to non-default values
+ get_training_widget.yaml_files.value = ["file_1.yml", "file_2.yml"]
+ get_training_widget.continue_training.value = True
+ get_training_widget.epochs.value = 50
+ get_training_widget.test_fraction.value = 0.20
+
+ # click reset button
+ get_training_widget.reset_button.clicked()
+
+ # check values have been reset
+ assert len(get_training_widget.yaml_files.value) == 1
+ assert get_training_widget.yaml_files.value[0] == Path.home()
+ assert not get_training_widget.continue_training.value
+ assert get_training_widget.epochs.value == 100
+ assert get_training_widget.test_fraction.value == 0.10
+
+
+def test_run_with_no_yaml_files(get_training_widget):
+ """
+ Checks whether expected info message will be shown to user if they don't
+ specify YAML file(s).
+ """
+ with patch("cellfinder.napari.train.train.show_info") as show_info:
+ get_training_widget.call_button.clicked()
+ show_info.assert_called_once_with(
+ "Please select a YAML file for training"
+ )
+
+
+def test_run_with_virtual_yaml_files(get_training_widget):
+ """
+ Checks that training is run with expected set of parameters.
+ """
+ with patch("cellfinder.napari.train.train.run_training") as run_training:
+ # make default input valid - need yml files (they don't technically
+ # have to exist)
+ virtual_yaml_files = (
+ Path.home() / "file_1.yml",
+ Path.home() / "file_2.yml",
+ )
+ get_training_widget.yaml_files.value = virtual_yaml_files
+ get_training_widget.call_button.clicked()
+
+ # create expected arguments for run
+ expected_training_args = TrainingDataInputs()
+ expected_network_args = OptionalNetworkInputs()
+ expected_optional_training_args = OptionalTrainingInputs()
+ expected_misc_args = MiscTrainingInputs()
+
+ # we expect the widget to make some changes to the defaults
+ # displayed before calling the training backend
+ expected_training_args.yaml_files = virtual_yaml_files
+ expected_network_args.trained_model = None
+ expected_network_args.model_weights = None
+
+ run_training.assert_called_once_with(
+ expected_training_args,
+ expected_network_args,
+ expected_optional_training_args,
+ expected_misc_args,
+ )
diff --git a/tests/napari/test_utils.py b/tests/napari/test_utils.py
new file mode 100644
index 00000000..480b5457
--- /dev/null
+++ b/tests/napari/test_utils.py
@@ -0,0 +1,64 @@
+import pytest
+from brainglobe_utils.cells.cells import Cell
+from qtpy.QtWidgets import QGridLayout
+
+from cellfinder.napari.utils import (
+ add_button,
+ add_combobox,
+ add_layers,
+ html_label_widget,
+)
+
+
+def test_add_layers(make_napari_viewer):
+ """Smoke test for add_layers utility"""
+ points = [
+ Cell(pos=[1, 2, 3], cell_type=Cell.CELL),
+ Cell(pos=[4, 5, 6], cell_type=Cell.UNKNOWN),
+ ]
+ viewer = make_napari_viewer()
+ n_layers = len(viewer.layers)
+ add_layers(points, viewer) # adds a "detected" and a "rejected layer"
+ assert len(viewer.layers) == n_layers + 2
+
+
+def test_html_label_widget():
+ """Simple unit test for the HTML Label widget"""
+ label_widget = html_label_widget("A nice label", tag="h1")
+ assert label_widget["widget_type"] == "Label"
+ assert label_widget["label"] == "A nice label
"
+
+
+@pytest.mark.parametrize("label_stack", [True, False])
+@pytest.mark.parametrize("label", ["A label", None])
+def test_add_combobox(label, label_stack):
+ """
+ Smoke test for add_combobox for all conditional branches
+ """
+ layout = QGridLayout()
+ combobox = add_combobox(
+ layout,
+ row=0,
+ label=label,
+ items=["item 1", "item 2"],
+ label_stack=label_stack,
+ )
+ assert combobox is not None
+
+
+@pytest.mark.parametrize(
+ argnames="alignment", argvalues=["center", "left", "right"]
+)
+def test_add_button(alignment):
+ """
+ Smoke tests for add_button for all conditional branches
+ """
+ layout = QGridLayout()
+ button = add_button(
+ layout=layout,
+ connected_function=lambda: None,
+ label="A button",
+ row=0,
+ alignment=alignment,
+ )
+ assert button is not None
diff --git a/tox.ini b/tox.ini
index 3f00afe3..a8505a09 100644
--- a/tox.ini
+++ b/tox.ini
@@ -1,6 +1,6 @@
# For more information about tox, see https://tox.readthedocs.io/en/latest/
[tox]
-envlist = py{38,39,310}
+envlist = py{38,39,310,311}
isolated_build = true
[gh-actions]
@@ -8,6 +8,7 @@ python =
3.8: py38
3.9: py39
3.10: py310
+ 3.11: py311
[testenv]
commands = python -m pytest -v --color=yes
@@ -17,5 +18,17 @@ deps =
pytest-lazy-fixture
pytest-mock
pytest-timeout
+ # Even though napari is a requirement for cellfinder-napari, we have to
+ # ensure it is installed with the default Qt backend here.
+ napari[all]
+ pytest-qt
+extras =
+ napari
passenv =
NUMBA_DISABLE_JIT
+ CI
+ GITHUB_ACTIONS
+ DISPLAY
+ XAUTHORITY
+ NUMPY_EXPERIMENTAL_ARRAY_FUNCTION
+ PYVISTA_OFF_SCREEN
+