diff --git a/.github/workflows/pypi.yml b/.github/workflows/pypi.yml
index eb99669..78f4d6f 100644
--- a/.github/workflows/pypi.yml
+++ b/.github/workflows/pypi.yml
@@ -36,7 +36,7 @@ jobs:
runs-on: ubuntu-latest
environment:
name: pypi
- url: https://pypi.org/p/cardioception
+ url: https://pypi.org/p/cardioception-toolbox
permissions:
id-token: write # IMPORTANT: mandatory for trusted publishing
steps:
diff --git a/README.md b/README.md
index f169e3b..e2c5f07 100644
--- a/README.md
+++ b/README.md
@@ -1,4 +1,4 @@
-[](https://github.com/LegrandNico/Cardioception/blob/master/LICENSE) [](https://GitHub.com/LegrandNico/Cardioception/releases/) [](https://github.com/pre-commit/pre-commit) [](https://badge.fury.io/py/cardioception) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/)
+[](https://github.com/LegrandNico/cardioception-toolbox/blob/master/LICENSE) [](https://github.com/LegrandNico/cardioception-toolbox/releases/) [](https://github.com/pre-commit/pre-commit) [](https://badge.fury.io/py/cardioception-toolbox) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/)
***
@@ -118,7 +118,7 @@ parameters['win'].close()
This minimal example will run the Heart Rate Discrimination task with a total of 10 trials using a Psi staircase.
-We provide standard scripts in the [wrappers](https://github.com/LegrandNico/Cardioception/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
+We provide standard scripts in the [wrappers](https://github.com/LegrandNico/cardioception-toolbox/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
## Creating a shortcut (Windows)
@@ -169,8 +169,8 @@ You can also analyze the results in [Google Colab](https://colab.research.google
| Notebook | Colab | nbViewer |
| --- | ---| --- |
-| Heartbeat Counting task report | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/cardioception/notebooks/HeartBeatCounting.ipynb) | [](https://nbviewer.jupyter.org/github/LegrandNico/Cardioception/blob/master/cardioception/notebooks/HeartBeatCounting.ipynb)
-| Heart Rate Discrimination task report | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/cardioception/notebooks/HeartRateDiscrimination.ipynb) | [](https://nbviewer.jupyter.org/github/LegrandNico/Cardioception/blob/master/cardioception/notebooks/HeartRateDiscrimination.ipynb)
+| Heartbeat Counting task report | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/cardioception/notebooks/HeartBeatCounting.ipynb) |
+| Heart Rate Discrimination task report | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/cardioception/notebooks/HeartRateDiscrimination.ipynb) |
## Bayesian modeling
@@ -178,8 +178,8 @@ More advanced subject and group-level Bayesian modelling approaches are describe
| Notebook | Colab | nbViewer |
| --- | ---| --- |
-| Fitting the psychometric function (single subject) | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb) | [](https://nbviewer.jupyter.org/github/LegrandNico/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
-| Fitting the psychometric function (group level) | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb) | [](https://nbviewer.jupyter.org/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
+| Fitting the psychometric function (single subject) | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb) |
+| Fitting the psychometric function (group level) | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb) |
# References
@@ -193,7 +193,7 @@ More advanced subject and group-level Bayesian modelling approaches are describe
This package is a fork of the original [Cardioception](https://github.com/embodied-computation-group/Cardioception) repository and is maintained by [Nicolas Legrand](https://github.com/LegrandNico).
- +
+ # Credit
diff --git a/docs/source/cite.md b/docs/source/cite.md
index 3d7a8a9..05d9de0 100644
--- a/docs/source/cite.md
+++ b/docs/source/cite.md
@@ -1,6 +1,6 @@
# How to cite?
-If you are using [Cardioception](https://github.com/LegrandNico/Cardioception) for your research, we ask you to cite the following paper in the final publication:
+If you are using the [cardioception toolbox](https://github.com/LegrandNico/cardioception-toolbox) for your research, we ask you to cite the following paper in the final publication:
* Legrand, N., Nikolova, N., Correa, C., Brændholt, M., Stuckert, A., Kildahl, N., Vejlø, M., Fardo, F., & Allen, M. (2021). The Heart Rate Discrimination Task: A psychophysical method to estimate the accuracy and precision of interoceptive beliefs. Biological Psychology, 108239.
diff --git a/docs/source/examples/R/Example scripts/Example_analysis_simple.Rmd b/docs/source/examples/R/Example scripts/Example_analysis_simple.Rmd
index fd0cd8c..c31102e 100644
--- a/docs/source/examples/R/Example scripts/Example_analysis_simple.Rmd
+++ b/docs/source/examples/R/Example scripts/Example_analysis_simple.Rmd
@@ -17,7 +17,7 @@ np <- import("numpy")
### **Reading in the data, here the working diectory has be where the exmaple scripts are located**
```{r message=FALSE}
#This line reads in a subject result file:
-df = read_csv("https://raw.githubusercontent.com/LegrandNico/Cardioception/master/docs/source/examples/templates/data/HRD/HRD_final.txt")
+df = read_csv("https://raw.githubusercontent.com/LegrandNico/cardioception-toolbox/master/docs/source/examples/templates/data/HRD/HRD_final.txt")
#These next lines read in the psi posterior distributions for each modality.
#These will be saved with names depending on your conditions, so modify as needed.
diff --git a/docs/source/index.md b/docs/source/index.md
index 7354892..8f5ba9d 100644
--- a/docs/source/index.md
+++ b/docs/source/index.md
@@ -1,10 +1,10 @@
# Cardioception toolbox
-[](https://github.com/LegrandNico/Cardioception/blob/master/LICENSE) [](https://GitHub.com/LegrandNico/Cardioception/releases/) [](https://github.com/pre-commit/pre-commit) [](https://badge.fury.io/py/cardioception) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/)
+[](https://github.com/LegrandNico/cardioception-toolbox/blob/master/LICENSE) [](https://github.com/LegrandNico/cardioception-toolbox/releases/) [](https://github.com/pre-commit/pre-commit) [](https://badge.fury.io/py/cardioception-toolbox) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/)
---
-
# Credit
diff --git a/docs/source/cite.md b/docs/source/cite.md
index 3d7a8a9..05d9de0 100644
--- a/docs/source/cite.md
+++ b/docs/source/cite.md
@@ -1,6 +1,6 @@
# How to cite?
-If you are using [Cardioception](https://github.com/LegrandNico/Cardioception) for your research, we ask you to cite the following paper in the final publication:
+If you are using the [cardioception toolbox](https://github.com/LegrandNico/cardioception-toolbox) for your research, we ask you to cite the following paper in the final publication:
* Legrand, N., Nikolova, N., Correa, C., Brændholt, M., Stuckert, A., Kildahl, N., Vejlø, M., Fardo, F., & Allen, M. (2021). The Heart Rate Discrimination Task: A psychophysical method to estimate the accuracy and precision of interoceptive beliefs. Biological Psychology, 108239.
diff --git a/docs/source/examples/R/Example scripts/Example_analysis_simple.Rmd b/docs/source/examples/R/Example scripts/Example_analysis_simple.Rmd
index fd0cd8c..c31102e 100644
--- a/docs/source/examples/R/Example scripts/Example_analysis_simple.Rmd
+++ b/docs/source/examples/R/Example scripts/Example_analysis_simple.Rmd
@@ -17,7 +17,7 @@ np <- import("numpy")
### **Reading in the data, here the working diectory has be where the exmaple scripts are located**
```{r message=FALSE}
#This line reads in a subject result file:
-df = read_csv("https://raw.githubusercontent.com/LegrandNico/Cardioception/master/docs/source/examples/templates/data/HRD/HRD_final.txt")
+df = read_csv("https://raw.githubusercontent.com/LegrandNico/cardioception-toolbox/master/docs/source/examples/templates/data/HRD/HRD_final.txt")
#These next lines read in the psi posterior distributions for each modality.
#These will be saved with names depending on your conditions, so modify as needed.
diff --git a/docs/source/index.md b/docs/source/index.md
index 7354892..8f5ba9d 100644
--- a/docs/source/index.md
+++ b/docs/source/index.md
@@ -1,10 +1,10 @@
# Cardioception toolbox
-[](https://github.com/LegrandNico/Cardioception/blob/master/LICENSE) [](https://GitHub.com/LegrandNico/Cardioception/releases/) [](https://github.com/pre-commit/pre-commit) [](https://badge.fury.io/py/cardioception) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/)
+[](https://github.com/LegrandNico/cardioception-toolbox/blob/master/LICENSE) [](https://github.com/LegrandNico/cardioception-toolbox/releases/) [](https://github.com/pre-commit/pre-commit) [](https://badge.fury.io/py/cardioception-toolbox) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/)
---
- +
+ ```{important}
The Cardioception Python Toolbox is a fork of the [original cardioception repository](https://github.com/embodied-computation-group/Cardioception) that I ([Nicolas Legrand](https://github.com/LegrandNico/)) created while working in [the ECG lab](https://www.the-ecg.org/) from 2019 to 2022. My previous lab has taken full control of the repository since then, meaning that I am unfortunately unable to maintain it as it should be. This repository allows me to pursue the maintenance of the package, aiming to provide reliable and robust tasks to measure cardiac interoception, together with computational modelling tools to analyse data gathered with these tasks.
@@ -26,7 +26,7 @@ These tasks can run using minimal experimental settings: a computer and a record
If you have questions regarding the tasks, want to report a bug or discuss data analysis, please ask on the public discussion page in this repository.
-If you want to report a bug, you can open an issue on the [GitHub page](https://github.com/LegrandNico/Cardioception).
+If you want to report a bug, you can open an issue on the [GitHub page](https://github.com/LegrandNico/cardioception-toolbox).
## Development
diff --git a/docs/source/measuring.md b/docs/source/measuring.md
index e19f5f2..1b281b3 100644
--- a/docs/source/measuring.md
+++ b/docs/source/measuring.md
@@ -1,14 +1,14 @@
# Measuring cardiac interoception
-Cardiac interoception has been largely investigated using the heartbeat counting task (also known as the heartbeat tracking task) that was formally introduced more than 40 years ago {cite:p}`1981:schandry`. This task comes with several variants that can concern task instruction, experimental design or the scores derived to measure cardiac interoceptive accuracy and metacognition. Here, we describe the heartbeat counting task together with the heart rate discrimination task, that was recently proposed {cite:p}`2022:legrand` and is also implemented in [cardioception](https://github.com/LegrandNico/Cardioception).
+Cardiac interoception has been largely investigated using the heartbeat counting task (also known as the heartbeat tracking task) that was formally introduced more than 40 years ago {cite:p}`1981:schandry`. This task comes with several variants that can concern task instruction, experimental design or the scores derived to measure cardiac interoceptive accuracy and metacognition. Here, we describe the heartbeat counting task together with the heart rate discrimination task, that was recently proposed {cite:p}`2022:legrand` and is also implemented in [cardioception](https://github.com/LegrandNico/cardioception-toolbox).
## The Heart Beat Counting task
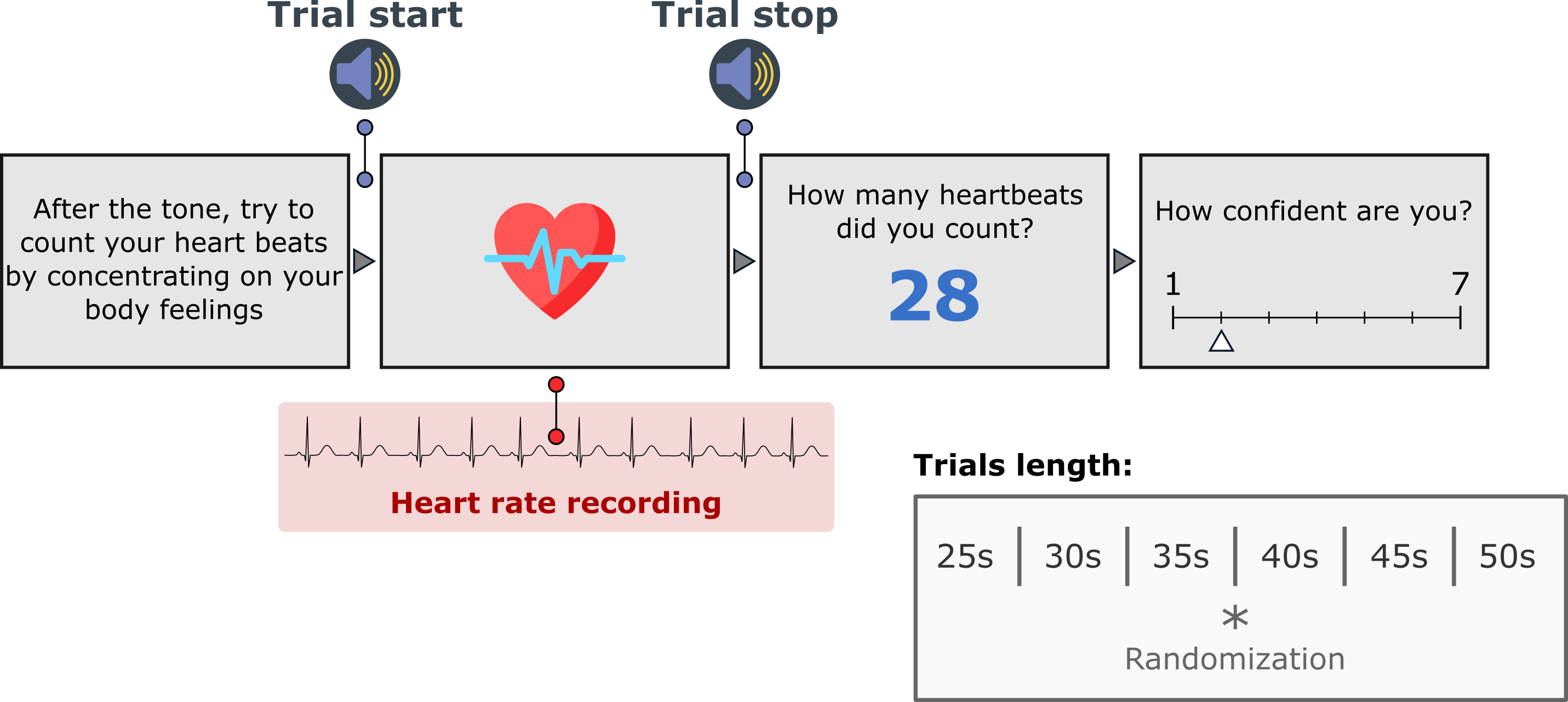
In the classic "heartbeat counting task" {cite:p}`1981:schandry,1978:dale` participants attend to their heartbeats in intervals of various lengths and are asked to count the number of heartbeats they can effectively feel during this period. An accuracy score is then derived by comparing the reported number of heartbeats and the true number of heartbeats. In the original version {cite:p}`1981:schandry`, the task started with a resting period of 60 seconds and consisted of three estimation sessions (25, 35, and 45 seconds) interleaved with resting periods of 30 seconds.
-
+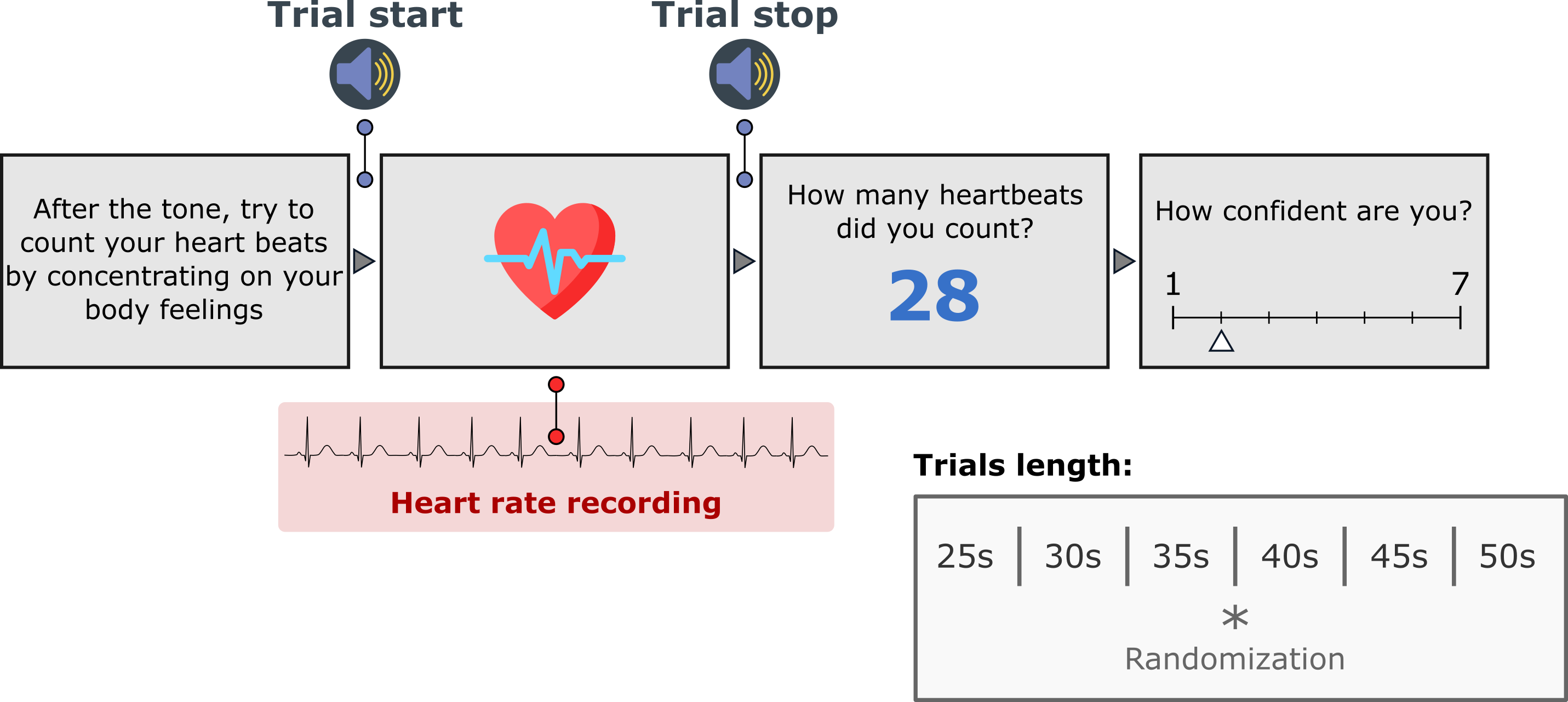
-By default, [Cardioception](https://github.com/LegrandNico/Cardioception) implements the version used in recent publications {cite:p}`2013:hart` in which a training trial of 20 seconds is proposed, after which the 6 experimental trials of different time windows (25, 30, 35,40, 45 and 50s) occurred in a randomized order. The trial length, the condition (`'Rest'`, `'Count'`, `'Training'`), and the randomization can be controlled in the parameters dictionary. This behaviour can be controlled using the `"taskVersion"` parameter.
+By default, [Cardioception](https://github.com/LegrandNico/cardioception-toolbox) implements the version used in recent publications {cite:p}`2013:hart` in which a training trial of 20 seconds is proposed, after which the 6 experimental trials of different time windows (25, 30, 35,40, 45 and 50s) occurred in a randomized order. The trial length, the condition (`'Rest'`, `'Count'`, `'Training'`), and the randomization can be controlled in the parameters dictionary. This behaviour can be controlled using the `"taskVersion"` parameter.
### Instructions
@@ -32,7 +32,7 @@ After each counting response, the participant is prompted to rate their subjecti
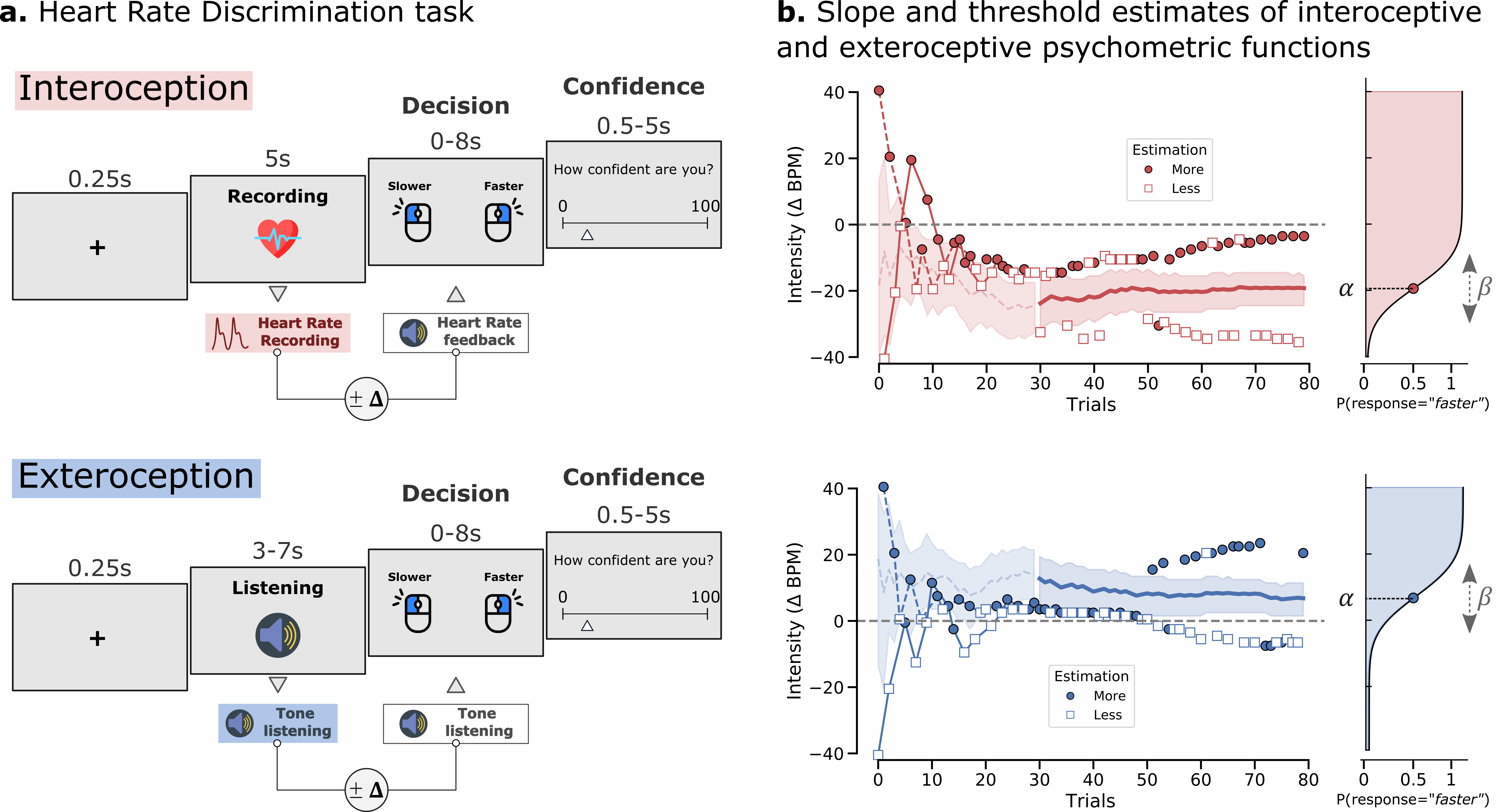
The **Heart Rate Discrimination Task** {cite:p}`2022:legrand` implements an adaptive psychophysical measure of cardiac interoception where participants have to estimate the frequency of their heart rate by comparing it to tones that can be faster or slower. By manipulating the difference between the true heart rate and the presented tone using different staircase procedures, the bias (threshold) and precision (slope) of the psychometric function can be estimated either online or offline, together with metacognitive efficiency.
-
+
### Staircases
diff --git a/docs/source/stats.md b/docs/source/stats.md
index 34aab82..5a2b10e 100644
--- a/docs/source/stats.md
+++ b/docs/source/stats.md
@@ -2,7 +2,7 @@
## Using R
-If you want to use R to analyse your data, you can find R/Stan scripts with example notebooks in [this folder](https://github.com/LegrandNico/Cardioception/tree/master/docs/source/examples/R).
+If you want to use R to analyse your data, you can find R/Stan scripts with example notebooks in [this folder](https://github.com/LegrandNico/cardioception-toolbox/tree/master/docs/source/examples/R).
## Using Python
@@ -61,8 +61,8 @@ examples/templates/*
| Notebook | Colab |
| --- | ---|
-| {ref}`hbc_template` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/templates/HeartBeatCounting.ipynb)
-| {ref}`hrd_template` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/templates/HeartRateDiscrimination.ipynb)
+| {ref}`hbc_template` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/templates/HeartBeatCounting.ipynb)
+| {ref}`hrd_template` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/templates/HeartRateDiscrimination.ipynb)
## Bayesian modelling of psychophysics
@@ -80,5 +80,5 @@ examples/psychophysics/*
| Notebook | Colab |
| --- | ---|
-| {ref}`psychophysics_subject_level` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
-| {ref}`psychophysics_group_level` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
+| {ref}`psychophysics_subject_level` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
+| {ref}`psychophysics_group_level` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
diff --git a/docs/source/user_guide.md b/docs/source/user_guide.md
index ac08179..4e969fb 100644
--- a/docs/source/user_guide.md
+++ b/docs/source/user_guide.md
@@ -90,7 +90,7 @@ parameters['win'].close()
This minimal example will run the Heart Rate Discrimination task with a total of 10 trials using a Psi staircase.
-We provide standard scripts in the [wrappers](https://github.com/LegrandNico/Cardioception/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
+We provide standard scripts in the [wrappers](https://github.com/LegrandNico/cardioception-toolbox/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
### Creating a shortcut (Windows)
diff --git a/setup.py b/setup.py
index faac0fc..ef7d77b 100644
--- a/setup.py
+++ b/setup.py
@@ -31,7 +31,7 @@ def get_version(rel_path):
LONG_DESCRIPTION = """Measuring cardiac interoception with Psychopy.
"""
-DISTNAME = "cardioception"
+DISTNAME = "cardioception-toolbox"
MAINTAINER = "Nicolas Legrand"
MAINTAINER_EMAIL = "nicolas.legrand@cas.au.dk"
diff --git a/tests/test_reports.py b/tests/test_reports.py
index 6f1991e..94ad4de 100644
--- a/tests/test_reports.py
+++ b/tests/test_reports.py
@@ -13,7 +13,7 @@ def test_preprocessing(self):
"""Test the preprocessing function"""
# load the main result data frame
results = pd.read_csv(
- "https://raw.githubusercontent.com/LegrandNico/Cardioception/master/docs/source/examples/templates/data/HRD/HRD_final.txt"
+ "https://raw.githubusercontent.com/LegrandNico/cardioception-toolbox/master/docs/source/examples/templates/data/HRD/HRD_final.txt"
)
preprocessing(results=results)
```{important}
The Cardioception Python Toolbox is a fork of the [original cardioception repository](https://github.com/embodied-computation-group/Cardioception) that I ([Nicolas Legrand](https://github.com/LegrandNico/)) created while working in [the ECG lab](https://www.the-ecg.org/) from 2019 to 2022. My previous lab has taken full control of the repository since then, meaning that I am unfortunately unable to maintain it as it should be. This repository allows me to pursue the maintenance of the package, aiming to provide reliable and robust tasks to measure cardiac interoception, together with computational modelling tools to analyse data gathered with these tasks.
@@ -26,7 +26,7 @@ These tasks can run using minimal experimental settings: a computer and a record
If you have questions regarding the tasks, want to report a bug or discuss data analysis, please ask on the public discussion page in this repository.
-If you want to report a bug, you can open an issue on the [GitHub page](https://github.com/LegrandNico/Cardioception).
+If you want to report a bug, you can open an issue on the [GitHub page](https://github.com/LegrandNico/cardioception-toolbox).
## Development
diff --git a/docs/source/measuring.md b/docs/source/measuring.md
index e19f5f2..1b281b3 100644
--- a/docs/source/measuring.md
+++ b/docs/source/measuring.md
@@ -1,14 +1,14 @@
# Measuring cardiac interoception
-Cardiac interoception has been largely investigated using the heartbeat counting task (also known as the heartbeat tracking task) that was formally introduced more than 40 years ago {cite:p}`1981:schandry`. This task comes with several variants that can concern task instruction, experimental design or the scores derived to measure cardiac interoceptive accuracy and metacognition. Here, we describe the heartbeat counting task together with the heart rate discrimination task, that was recently proposed {cite:p}`2022:legrand` and is also implemented in [cardioception](https://github.com/LegrandNico/Cardioception).
+Cardiac interoception has been largely investigated using the heartbeat counting task (also known as the heartbeat tracking task) that was formally introduced more than 40 years ago {cite:p}`1981:schandry`. This task comes with several variants that can concern task instruction, experimental design or the scores derived to measure cardiac interoceptive accuracy and metacognition. Here, we describe the heartbeat counting task together with the heart rate discrimination task, that was recently proposed {cite:p}`2022:legrand` and is also implemented in [cardioception](https://github.com/LegrandNico/cardioception-toolbox).
## The Heart Beat Counting task
In the classic "heartbeat counting task" {cite:p}`1981:schandry,1978:dale` participants attend to their heartbeats in intervals of various lengths and are asked to count the number of heartbeats they can effectively feel during this period. An accuracy score is then derived by comparing the reported number of heartbeats and the true number of heartbeats. In the original version {cite:p}`1981:schandry`, the task started with a resting period of 60 seconds and consisted of three estimation sessions (25, 35, and 45 seconds) interleaved with resting periods of 30 seconds.
-
+
-By default, [Cardioception](https://github.com/LegrandNico/Cardioception) implements the version used in recent publications {cite:p}`2013:hart` in which a training trial of 20 seconds is proposed, after which the 6 experimental trials of different time windows (25, 30, 35,40, 45 and 50s) occurred in a randomized order. The trial length, the condition (`'Rest'`, `'Count'`, `'Training'`), and the randomization can be controlled in the parameters dictionary. This behaviour can be controlled using the `"taskVersion"` parameter.
+By default, [Cardioception](https://github.com/LegrandNico/cardioception-toolbox) implements the version used in recent publications {cite:p}`2013:hart` in which a training trial of 20 seconds is proposed, after which the 6 experimental trials of different time windows (25, 30, 35,40, 45 and 50s) occurred in a randomized order. The trial length, the condition (`'Rest'`, `'Count'`, `'Training'`), and the randomization can be controlled in the parameters dictionary. This behaviour can be controlled using the `"taskVersion"` parameter.
### Instructions
@@ -32,7 +32,7 @@ After each counting response, the participant is prompted to rate their subjecti
The **Heart Rate Discrimination Task** {cite:p}`2022:legrand` implements an adaptive psychophysical measure of cardiac interoception where participants have to estimate the frequency of their heart rate by comparing it to tones that can be faster or slower. By manipulating the difference between the true heart rate and the presented tone using different staircase procedures, the bias (threshold) and precision (slope) of the psychometric function can be estimated either online or offline, together with metacognitive efficiency.
-
+
### Staircases
diff --git a/docs/source/stats.md b/docs/source/stats.md
index 34aab82..5a2b10e 100644
--- a/docs/source/stats.md
+++ b/docs/source/stats.md
@@ -2,7 +2,7 @@
## Using R
-If you want to use R to analyse your data, you can find R/Stan scripts with example notebooks in [this folder](https://github.com/LegrandNico/Cardioception/tree/master/docs/source/examples/R).
+If you want to use R to analyse your data, you can find R/Stan scripts with example notebooks in [this folder](https://github.com/LegrandNico/cardioception-toolbox/tree/master/docs/source/examples/R).
## Using Python
@@ -61,8 +61,8 @@ examples/templates/*
| Notebook | Colab |
| --- | ---|
-| {ref}`hbc_template` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/templates/HeartBeatCounting.ipynb)
-| {ref}`hrd_template` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/templates/HeartRateDiscrimination.ipynb)
+| {ref}`hbc_template` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/templates/HeartBeatCounting.ipynb)
+| {ref}`hrd_template` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/templates/HeartRateDiscrimination.ipynb)
## Bayesian modelling of psychophysics
@@ -80,5 +80,5 @@ examples/psychophysics/*
| Notebook | Colab |
| --- | ---|
-| {ref}`psychophysics_subject_level` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
-| {ref}`psychophysics_group_level` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
+| {ref}`psychophysics_subject_level` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
+| {ref}`psychophysics_group_level` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
diff --git a/docs/source/user_guide.md b/docs/source/user_guide.md
index ac08179..4e969fb 100644
--- a/docs/source/user_guide.md
+++ b/docs/source/user_guide.md
@@ -90,7 +90,7 @@ parameters['win'].close()
This minimal example will run the Heart Rate Discrimination task with a total of 10 trials using a Psi staircase.
-We provide standard scripts in the [wrappers](https://github.com/LegrandNico/Cardioception/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
+We provide standard scripts in the [wrappers](https://github.com/LegrandNico/cardioception-toolbox/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
### Creating a shortcut (Windows)
diff --git a/setup.py b/setup.py
index faac0fc..ef7d77b 100644
--- a/setup.py
+++ b/setup.py
@@ -31,7 +31,7 @@ def get_version(rel_path):
LONG_DESCRIPTION = """Measuring cardiac interoception with Psychopy.
"""
-DISTNAME = "cardioception"
+DISTNAME = "cardioception-toolbox"
MAINTAINER = "Nicolas Legrand"
MAINTAINER_EMAIL = "nicolas.legrand@cas.au.dk"
diff --git a/tests/test_reports.py b/tests/test_reports.py
index 6f1991e..94ad4de 100644
--- a/tests/test_reports.py
+++ b/tests/test_reports.py
@@ -13,7 +13,7 @@ def test_preprocessing(self):
"""Test the preprocessing function"""
# load the main result data frame
results = pd.read_csv(
- "https://raw.githubusercontent.com/LegrandNico/Cardioception/master/docs/source/examples/templates/data/HRD/HRD_final.txt"
+ "https://raw.githubusercontent.com/LegrandNico/cardioception-toolbox/master/docs/source/examples/templates/data/HRD/HRD_final.txt"
)
preprocessing(results=results)
 +
+ ```{important}
The Cardioception Python Toolbox is a fork of the [original cardioception repository](https://github.com/embodied-computation-group/Cardioception) that I ([Nicolas Legrand](https://github.com/LegrandNico/)) created while working in [the ECG lab](https://www.the-ecg.org/) from 2019 to 2022. My previous lab has taken full control of the repository since then, meaning that I am unfortunately unable to maintain it as it should be. This repository allows me to pursue the maintenance of the package, aiming to provide reliable and robust tasks to measure cardiac interoception, together with computational modelling tools to analyse data gathered with these tasks.
@@ -26,7 +26,7 @@ These tasks can run using minimal experimental settings: a computer and a record
If you have questions regarding the tasks, want to report a bug or discuss data analysis, please ask on the public discussion page in this repository.
-If you want to report a bug, you can open an issue on the [GitHub page](https://github.com/LegrandNico/Cardioception).
+If you want to report a bug, you can open an issue on the [GitHub page](https://github.com/LegrandNico/cardioception-toolbox).
## Development
diff --git a/docs/source/measuring.md b/docs/source/measuring.md
index e19f5f2..1b281b3 100644
--- a/docs/source/measuring.md
+++ b/docs/source/measuring.md
@@ -1,14 +1,14 @@
# Measuring cardiac interoception
-Cardiac interoception has been largely investigated using the heartbeat counting task (also known as the heartbeat tracking task) that was formally introduced more than 40 years ago {cite:p}`1981:schandry`. This task comes with several variants that can concern task instruction, experimental design or the scores derived to measure cardiac interoceptive accuracy and metacognition. Here, we describe the heartbeat counting task together with the heart rate discrimination task, that was recently proposed {cite:p}`2022:legrand` and is also implemented in [cardioception](https://github.com/LegrandNico/Cardioception).
+Cardiac interoception has been largely investigated using the heartbeat counting task (also known as the heartbeat tracking task) that was formally introduced more than 40 years ago {cite:p}`1981:schandry`. This task comes with several variants that can concern task instruction, experimental design or the scores derived to measure cardiac interoceptive accuracy and metacognition. Here, we describe the heartbeat counting task together with the heart rate discrimination task, that was recently proposed {cite:p}`2022:legrand` and is also implemented in [cardioception](https://github.com/LegrandNico/cardioception-toolbox).
## The Heart Beat Counting task
In the classic "heartbeat counting task" {cite:p}`1981:schandry,1978:dale` participants attend to their heartbeats in intervals of various lengths and are asked to count the number of heartbeats they can effectively feel during this period. An accuracy score is then derived by comparing the reported number of heartbeats and the true number of heartbeats. In the original version {cite:p}`1981:schandry`, the task started with a resting period of 60 seconds and consisted of three estimation sessions (25, 35, and 45 seconds) interleaved with resting periods of 30 seconds.
-
+
-By default, [Cardioception](https://github.com/LegrandNico/Cardioception) implements the version used in recent publications {cite:p}`2013:hart` in which a training trial of 20 seconds is proposed, after which the 6 experimental trials of different time windows (25, 30, 35,40, 45 and 50s) occurred in a randomized order. The trial length, the condition (`'Rest'`, `'Count'`, `'Training'`), and the randomization can be controlled in the parameters dictionary. This behaviour can be controlled using the `"taskVersion"` parameter.
+By default, [Cardioception](https://github.com/LegrandNico/cardioception-toolbox) implements the version used in recent publications {cite:p}`2013:hart` in which a training trial of 20 seconds is proposed, after which the 6 experimental trials of different time windows (25, 30, 35,40, 45 and 50s) occurred in a randomized order. The trial length, the condition (`'Rest'`, `'Count'`, `'Training'`), and the randomization can be controlled in the parameters dictionary. This behaviour can be controlled using the `"taskVersion"` parameter.
### Instructions
@@ -32,7 +32,7 @@ After each counting response, the participant is prompted to rate their subjecti
The **Heart Rate Discrimination Task** {cite:p}`2022:legrand` implements an adaptive psychophysical measure of cardiac interoception where participants have to estimate the frequency of their heart rate by comparing it to tones that can be faster or slower. By manipulating the difference between the true heart rate and the presented tone using different staircase procedures, the bias (threshold) and precision (slope) of the psychometric function can be estimated either online or offline, together with metacognitive efficiency.
-
+
### Staircases
diff --git a/docs/source/stats.md b/docs/source/stats.md
index 34aab82..5a2b10e 100644
--- a/docs/source/stats.md
+++ b/docs/source/stats.md
@@ -2,7 +2,7 @@
## Using R
-If you want to use R to analyse your data, you can find R/Stan scripts with example notebooks in [this folder](https://github.com/LegrandNico/Cardioception/tree/master/docs/source/examples/R).
+If you want to use R to analyse your data, you can find R/Stan scripts with example notebooks in [this folder](https://github.com/LegrandNico/cardioception-toolbox/tree/master/docs/source/examples/R).
## Using Python
@@ -61,8 +61,8 @@ examples/templates/*
| Notebook | Colab |
| --- | ---|
-| {ref}`hbc_template` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/templates/HeartBeatCounting.ipynb)
-| {ref}`hrd_template` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/templates/HeartRateDiscrimination.ipynb)
+| {ref}`hbc_template` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/templates/HeartBeatCounting.ipynb)
+| {ref}`hrd_template` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/templates/HeartRateDiscrimination.ipynb)
## Bayesian modelling of psychophysics
@@ -80,5 +80,5 @@ examples/psychophysics/*
| Notebook | Colab |
| --- | ---|
-| {ref}`psychophysics_subject_level` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
-| {ref}`psychophysics_group_level` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
+| {ref}`psychophysics_subject_level` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
+| {ref}`psychophysics_group_level` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
diff --git a/docs/source/user_guide.md b/docs/source/user_guide.md
index ac08179..4e969fb 100644
--- a/docs/source/user_guide.md
+++ b/docs/source/user_guide.md
@@ -90,7 +90,7 @@ parameters['win'].close()
This minimal example will run the Heart Rate Discrimination task with a total of 10 trials using a Psi staircase.
-We provide standard scripts in the [wrappers](https://github.com/LegrandNico/Cardioception/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
+We provide standard scripts in the [wrappers](https://github.com/LegrandNico/cardioception-toolbox/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
### Creating a shortcut (Windows)
diff --git a/setup.py b/setup.py
index faac0fc..ef7d77b 100644
--- a/setup.py
+++ b/setup.py
@@ -31,7 +31,7 @@ def get_version(rel_path):
LONG_DESCRIPTION = """Measuring cardiac interoception with Psychopy.
"""
-DISTNAME = "cardioception"
+DISTNAME = "cardioception-toolbox"
MAINTAINER = "Nicolas Legrand"
MAINTAINER_EMAIL = "nicolas.legrand@cas.au.dk"
diff --git a/tests/test_reports.py b/tests/test_reports.py
index 6f1991e..94ad4de 100644
--- a/tests/test_reports.py
+++ b/tests/test_reports.py
@@ -13,7 +13,7 @@ def test_preprocessing(self):
"""Test the preprocessing function"""
# load the main result data frame
results = pd.read_csv(
- "https://raw.githubusercontent.com/LegrandNico/Cardioception/master/docs/source/examples/templates/data/HRD/HRD_final.txt"
+ "https://raw.githubusercontent.com/LegrandNico/cardioception-toolbox/master/docs/source/examples/templates/data/HRD/HRD_final.txt"
)
preprocessing(results=results)
```{important}
The Cardioception Python Toolbox is a fork of the [original cardioception repository](https://github.com/embodied-computation-group/Cardioception) that I ([Nicolas Legrand](https://github.com/LegrandNico/)) created while working in [the ECG lab](https://www.the-ecg.org/) from 2019 to 2022. My previous lab has taken full control of the repository since then, meaning that I am unfortunately unable to maintain it as it should be. This repository allows me to pursue the maintenance of the package, aiming to provide reliable and robust tasks to measure cardiac interoception, together with computational modelling tools to analyse data gathered with these tasks.
@@ -26,7 +26,7 @@ These tasks can run using minimal experimental settings: a computer and a record
If you have questions regarding the tasks, want to report a bug or discuss data analysis, please ask on the public discussion page in this repository.
-If you want to report a bug, you can open an issue on the [GitHub page](https://github.com/LegrandNico/Cardioception).
+If you want to report a bug, you can open an issue on the [GitHub page](https://github.com/LegrandNico/cardioception-toolbox).
## Development
diff --git a/docs/source/measuring.md b/docs/source/measuring.md
index e19f5f2..1b281b3 100644
--- a/docs/source/measuring.md
+++ b/docs/source/measuring.md
@@ -1,14 +1,14 @@
# Measuring cardiac interoception
-Cardiac interoception has been largely investigated using the heartbeat counting task (also known as the heartbeat tracking task) that was formally introduced more than 40 years ago {cite:p}`1981:schandry`. This task comes with several variants that can concern task instruction, experimental design or the scores derived to measure cardiac interoceptive accuracy and metacognition. Here, we describe the heartbeat counting task together with the heart rate discrimination task, that was recently proposed {cite:p}`2022:legrand` and is also implemented in [cardioception](https://github.com/LegrandNico/Cardioception).
+Cardiac interoception has been largely investigated using the heartbeat counting task (also known as the heartbeat tracking task) that was formally introduced more than 40 years ago {cite:p}`1981:schandry`. This task comes with several variants that can concern task instruction, experimental design or the scores derived to measure cardiac interoceptive accuracy and metacognition. Here, we describe the heartbeat counting task together with the heart rate discrimination task, that was recently proposed {cite:p}`2022:legrand` and is also implemented in [cardioception](https://github.com/LegrandNico/cardioception-toolbox).
## The Heart Beat Counting task
In the classic "heartbeat counting task" {cite:p}`1981:schandry,1978:dale` participants attend to their heartbeats in intervals of various lengths and are asked to count the number of heartbeats they can effectively feel during this period. An accuracy score is then derived by comparing the reported number of heartbeats and the true number of heartbeats. In the original version {cite:p}`1981:schandry`, the task started with a resting period of 60 seconds and consisted of three estimation sessions (25, 35, and 45 seconds) interleaved with resting periods of 30 seconds.
-
+
-By default, [Cardioception](https://github.com/LegrandNico/Cardioception) implements the version used in recent publications {cite:p}`2013:hart` in which a training trial of 20 seconds is proposed, after which the 6 experimental trials of different time windows (25, 30, 35,40, 45 and 50s) occurred in a randomized order. The trial length, the condition (`'Rest'`, `'Count'`, `'Training'`), and the randomization can be controlled in the parameters dictionary. This behaviour can be controlled using the `"taskVersion"` parameter.
+By default, [Cardioception](https://github.com/LegrandNico/cardioception-toolbox) implements the version used in recent publications {cite:p}`2013:hart` in which a training trial of 20 seconds is proposed, after which the 6 experimental trials of different time windows (25, 30, 35,40, 45 and 50s) occurred in a randomized order. The trial length, the condition (`'Rest'`, `'Count'`, `'Training'`), and the randomization can be controlled in the parameters dictionary. This behaviour can be controlled using the `"taskVersion"` parameter.
### Instructions
@@ -32,7 +32,7 @@ After each counting response, the participant is prompted to rate their subjecti
The **Heart Rate Discrimination Task** {cite:p}`2022:legrand` implements an adaptive psychophysical measure of cardiac interoception where participants have to estimate the frequency of their heart rate by comparing it to tones that can be faster or slower. By manipulating the difference between the true heart rate and the presented tone using different staircase procedures, the bias (threshold) and precision (slope) of the psychometric function can be estimated either online or offline, together with metacognitive efficiency.
-
+
### Staircases
diff --git a/docs/source/stats.md b/docs/source/stats.md
index 34aab82..5a2b10e 100644
--- a/docs/source/stats.md
+++ b/docs/source/stats.md
@@ -2,7 +2,7 @@
## Using R
-If you want to use R to analyse your data, you can find R/Stan scripts with example notebooks in [this folder](https://github.com/LegrandNico/Cardioception/tree/master/docs/source/examples/R).
+If you want to use R to analyse your data, you can find R/Stan scripts with example notebooks in [this folder](https://github.com/LegrandNico/cardioception-toolbox/tree/master/docs/source/examples/R).
## Using Python
@@ -61,8 +61,8 @@ examples/templates/*
| Notebook | Colab |
| --- | ---|
-| {ref}`hbc_template` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/templates/HeartBeatCounting.ipynb)
-| {ref}`hrd_template` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/templates/HeartRateDiscrimination.ipynb)
+| {ref}`hbc_template` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/templates/HeartBeatCounting.ipynb)
+| {ref}`hrd_template` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/templates/HeartRateDiscrimination.ipynb)
## Bayesian modelling of psychophysics
@@ -80,5 +80,5 @@ examples/psychophysics/*
| Notebook | Colab |
| --- | ---|
-| {ref}`psychophysics_subject_level` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
-| {ref}`psychophysics_group_level` | [](https://colab.research.google.com/github/LegrandNico/Cardioception/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
+| {ref}`psychophysics_subject_level` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/1-psychophysics_subject_level.ipynb)
+| {ref}`psychophysics_group_level` | [](https://colab.research.google.com/github/LegrandNico/cardioception-toolbox/blob/master/docs/source/examples/psychophysics/2-psychophysics_group_level.ipynb)
diff --git a/docs/source/user_guide.md b/docs/source/user_guide.md
index ac08179..4e969fb 100644
--- a/docs/source/user_guide.md
+++ b/docs/source/user_guide.md
@@ -90,7 +90,7 @@ parameters['win'].close()
This minimal example will run the Heart Rate Discrimination task with a total of 10 trials using a Psi staircase.
-We provide standard scripts in the [wrappers](https://github.com/LegrandNico/Cardioception/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
+We provide standard scripts in the [wrappers](https://github.com/LegrandNico/cardioception-toolbox/tree/master/wrappers) folder that can be adapted to your needs. We recommend copying this script in your local task folder if you want to parametrize it to fit your needs. The tasks can then easily be executed by running the corresponding wrapper file (e.g. in a terminal).
### Creating a shortcut (Windows)
diff --git a/setup.py b/setup.py
index faac0fc..ef7d77b 100644
--- a/setup.py
+++ b/setup.py
@@ -31,7 +31,7 @@ def get_version(rel_path):
LONG_DESCRIPTION = """Measuring cardiac interoception with Psychopy.
"""
-DISTNAME = "cardioception"
+DISTNAME = "cardioception-toolbox"
MAINTAINER = "Nicolas Legrand"
MAINTAINER_EMAIL = "nicolas.legrand@cas.au.dk"
diff --git a/tests/test_reports.py b/tests/test_reports.py
index 6f1991e..94ad4de 100644
--- a/tests/test_reports.py
+++ b/tests/test_reports.py
@@ -13,7 +13,7 @@ def test_preprocessing(self):
"""Test the preprocessing function"""
# load the main result data frame
results = pd.read_csv(
- "https://raw.githubusercontent.com/LegrandNico/Cardioception/master/docs/source/examples/templates/data/HRD/HRD_final.txt"
+ "https://raw.githubusercontent.com/LegrandNico/cardioception-toolbox/master/docs/source/examples/templates/data/HRD/HRD_final.txt"
)
preprocessing(results=results)
 +
+ # Credit
diff --git a/docs/source/cite.md b/docs/source/cite.md
index 3d7a8a9..05d9de0 100644
--- a/docs/source/cite.md
+++ b/docs/source/cite.md
@@ -1,6 +1,6 @@
# How to cite?
-If you are using [Cardioception](https://github.com/LegrandNico/Cardioception) for your research, we ask you to cite the following paper in the final publication:
+If you are using the [cardioception toolbox](https://github.com/LegrandNico/cardioception-toolbox) for your research, we ask you to cite the following paper in the final publication:
* Legrand, N., Nikolova, N., Correa, C., Brændholt, M., Stuckert, A., Kildahl, N., Vejlø, M., Fardo, F., & Allen, M. (2021). The Heart Rate Discrimination Task: A psychophysical method to estimate the accuracy and precision of interoceptive beliefs. Biological Psychology, 108239.
# Credit
diff --git a/docs/source/cite.md b/docs/source/cite.md
index 3d7a8a9..05d9de0 100644
--- a/docs/source/cite.md
+++ b/docs/source/cite.md
@@ -1,6 +1,6 @@
# How to cite?
-If you are using [Cardioception](https://github.com/LegrandNico/Cardioception) for your research, we ask you to cite the following paper in the final publication:
+If you are using the [cardioception toolbox](https://github.com/LegrandNico/cardioception-toolbox) for your research, we ask you to cite the following paper in the final publication:
* Legrand, N., Nikolova, N., Correa, C., Brændholt, M., Stuckert, A., Kildahl, N., Vejlø, M., Fardo, F., & Allen, M. (2021). The Heart Rate Discrimination Task: A psychophysical method to estimate the accuracy and precision of interoceptive beliefs. Biological Psychology, 108239.