diff --git a/.github/workflows/build.yml b/.github/workflows/build.yml
new file mode 100644
index 0000000..d710f21
--- /dev/null
+++ b/.github/workflows/build.yml
@@ -0,0 +1,31 @@
+name: build
+
+on: [push]
+
+jobs:
+ build:
+ runs-on: ubuntu-latest
+
+ steps:
+ - uses: actions/checkout@v2
+ - name: Set up Python 3.9
+ uses: actions/setup-python@v2
+ with:
+ python-version: "3.9"
+ - name: Install dependencies
+ run: |
+ python -m pip install --upgrade pip
+ pip install pytest pytest-cov coveralls
+ sudo apt-get -qq update
+ sudo apt-get install ncbi-blast+
+ - name: Install
+ run: |
+ pip install -e .
+ - name: Test with pytest
+ run: |
+ python -m pytest --cov geneblocks --cov-report term-missing
+ - name: Coveralls
+ run: coveralls
+ env:
+ GITHUB_TOKEN: ${{ secrets.GITHUB_TOKEN }}
+ COVERALLS_SERVICE_NAME: github
diff --git a/.travis.yml b/.travis.yml
index 6f4f4a5..49375c3 100644
--- a/.travis.yml
+++ b/.travis.yml
@@ -1,12 +1,12 @@
language: python
python:
- - "3.6"
+ - "3.9"
# command to install dependencies
before_install:
- sudo apt-get -qq update
- sudo apt-get install ncbi-blast+
install:
- - pip install coveralls pytest-cov==2.6 pytest==3.2.3
+ - pip install coveralls pytest-cov pytest
- pip install -e .
# command to run tests
script:
diff --git a/README.rst b/README.rst
index c2db6ad..aafee9d 100644
--- a/README.rst
+++ b/README.rst
@@ -8,9 +8,9 @@
GeneBlocks
==========
-.. image:: https://travis-ci.org/Edinburgh-Genome-Foundry/Geneblocks.svg?branch=master
- :target: https://travis-ci.org/Edinburgh-Genome-Foundry/Geneblocks
- :alt: Travis CI build status
+.. image:: https://github.com/Edinburgh-Genome-Foundry/Geneblocks/actions/workflows/build.yml/badge.svg
+ :target: https://github.com/Edinburgh-Genome-Foundry/Geneblocks/actions/workflows/build.yml
+ :alt: GitHub CI build status
.. image:: https://coveralls.io/repos/github/Edinburgh-Genome-Foundry/Geneblocks/badge.svg
:target: https://coveralls.io/github/Edinburgh-Genome-Foundry/Geneblocks
@@ -40,20 +40,20 @@ The CommonBlocks feature requires NCBI BLAST+. On Ubuntu, install it with
.. code:: shell
- (sudo) apt-get install ncbi-blast+
+ apt-get install ncbi-blast+
You can install GeneBlocks through PIP:
.. code:: shell
- (sudo) pip install geneblocks
+ pip install geneblocks
Alternatively, you can unzip the sources in a folder and type:
.. code:: shell
- (sudo) python setup.py install
+ python setup.py install
Usage
@@ -180,10 +180,10 @@ of modifications brought to the second sequence:
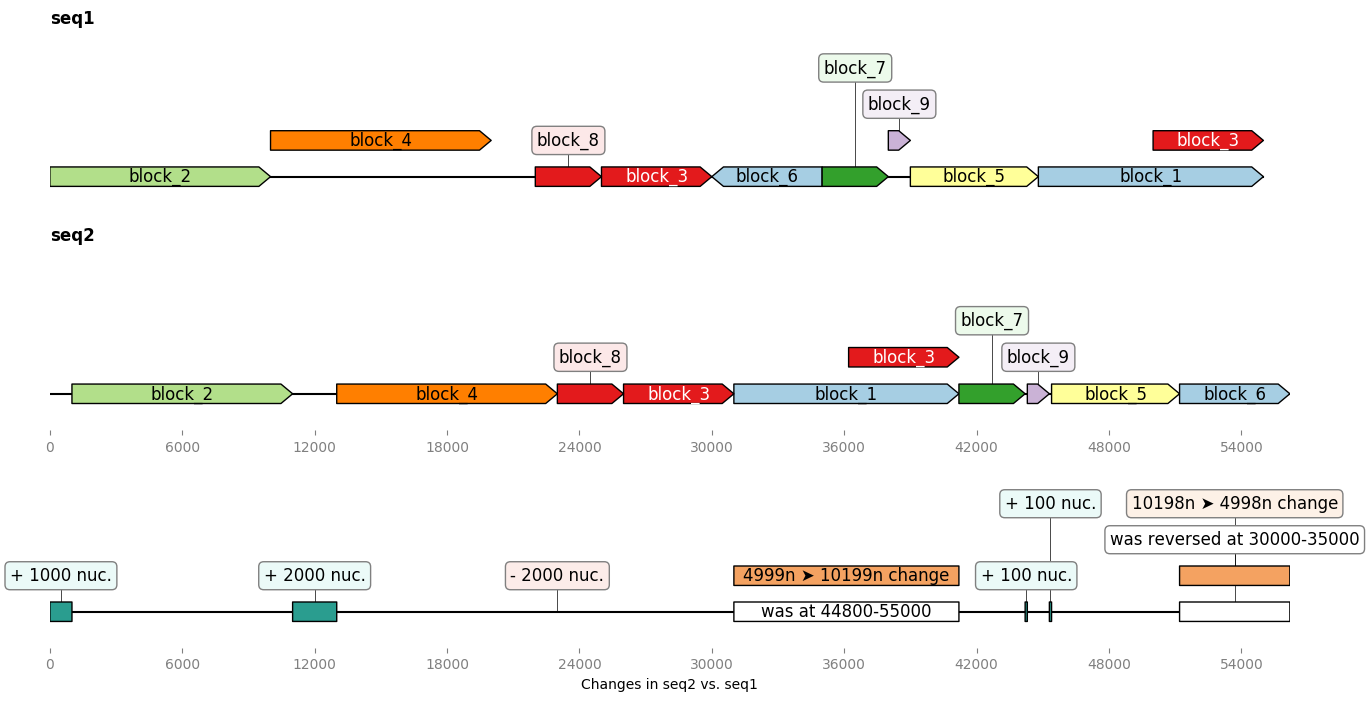 -Licence
--------
+License = MIT
+-------------
Geneblocks is an open-source software originally written at the `Edinburgh Genome Foundry
`_ by `Zulko `_
-and `released on Github `_ under the MIT licence (Copyright 2017 Edinburgh Genome Foundry).
+and `released on Github `_ under the MIT license (Copyright 2017 Edinburgh Genome Foundry).
Everyone is welcome to contribute!
diff --git a/pypi-readme.rst b/pypi-readme.rst
index 6b51784..b4eb8e0 100644
--- a/pypi-readme.rst
+++ b/pypi-readme.rst
@@ -1,9 +1,9 @@
GeneBlocks
-=============
+==========
-.. image:: https://travis-ci.org/Edinburgh-Genome-Foundry/DnaChisel.svg?branch=master
- :target: https://travis-ci.org/Edinburgh-Genome-Foundry/Geneblocks
- :alt: Travis CI build status
+.. image:: https://github.com/Edinburgh-Genome-Foundry/Geneblocks/actions/workflows/build.yml/badge.svg
+ :target: https://github.com/Edinburgh-Genome-Foundry/Geneblocks/actions/workflows/build.yml
+ :alt: GitHub CI build status
.. image:: https://coveralls.io/repos/github/Edinburgh-Genome-Foundry/Geneblocks/badge.svg
:target: https://coveralls.io/github/Edinburgh-Genome-Foundry/Geneblocks
diff --git a/setup.py b/setup.py
index 8f8abd8..defcdd6 100644
--- a/setup.py
+++ b/setup.py
@@ -1,19 +1,26 @@
import ez_setup
+
ez_setup.use_setuptools()
from setuptools import setup, find_packages
-exec(open('geneblocks/version.py').read()) # loads __version__
+exec(open("geneblocks/version.py").read()) # loads __version__
setup(
- name='geneblocks',
+ name="geneblocks",
version=__version__,
- author='Zulko',
- description='Library to compare DNA sequences (diff, common blocks, etc.)',
- long_description=open('pypi-readme.rst').read(),
- license='MIT',
- url='https://github.com/Edinburgh-Genome-Foundry/geneblocks',
+ author="Zulko",
+ description="Library to compare DNA sequences (diff, common blocks, etc.)",
+ long_description=open("pypi-readme.rst").read(),
+ license="MIT",
+ url="https://github.com/Edinburgh-Genome-Foundry/geneblocks",
keywords="DNA sequence blocks diff synthetic-biology bioinformatics",
- packages=find_packages(exclude='docs'),
- install_requires=['numpy', 'Biopython', 'dna_features_viewer',
- 'networkx', 'python-Levenshtein'])
+ packages=find_packages(exclude="docs"),
+ install_requires=[
+ "numpy",
+ "Biopython",
+ "dna_features_viewer",
+ "networkx",
+ "python-Levenshtein",
+ ],
+)
-Licence
--------
+License = MIT
+-------------
Geneblocks is an open-source software originally written at the `Edinburgh Genome Foundry
`_ by `Zulko `_
-and `released on Github `_ under the MIT licence (Copyright 2017 Edinburgh Genome Foundry).
+and `released on Github `_ under the MIT license (Copyright 2017 Edinburgh Genome Foundry).
Everyone is welcome to contribute!
diff --git a/pypi-readme.rst b/pypi-readme.rst
index 6b51784..b4eb8e0 100644
--- a/pypi-readme.rst
+++ b/pypi-readme.rst
@@ -1,9 +1,9 @@
GeneBlocks
-=============
+==========
-.. image:: https://travis-ci.org/Edinburgh-Genome-Foundry/DnaChisel.svg?branch=master
- :target: https://travis-ci.org/Edinburgh-Genome-Foundry/Geneblocks
- :alt: Travis CI build status
+.. image:: https://github.com/Edinburgh-Genome-Foundry/Geneblocks/actions/workflows/build.yml/badge.svg
+ :target: https://github.com/Edinburgh-Genome-Foundry/Geneblocks/actions/workflows/build.yml
+ :alt: GitHub CI build status
.. image:: https://coveralls.io/repos/github/Edinburgh-Genome-Foundry/Geneblocks/badge.svg
:target: https://coveralls.io/github/Edinburgh-Genome-Foundry/Geneblocks
diff --git a/setup.py b/setup.py
index 8f8abd8..defcdd6 100644
--- a/setup.py
+++ b/setup.py
@@ -1,19 +1,26 @@
import ez_setup
+
ez_setup.use_setuptools()
from setuptools import setup, find_packages
-exec(open('geneblocks/version.py').read()) # loads __version__
+exec(open("geneblocks/version.py").read()) # loads __version__
setup(
- name='geneblocks',
+ name="geneblocks",
version=__version__,
- author='Zulko',
- description='Library to compare DNA sequences (diff, common blocks, etc.)',
- long_description=open('pypi-readme.rst').read(),
- license='MIT',
- url='https://github.com/Edinburgh-Genome-Foundry/geneblocks',
+ author="Zulko",
+ description="Library to compare DNA sequences (diff, common blocks, etc.)",
+ long_description=open("pypi-readme.rst").read(),
+ license="MIT",
+ url="https://github.com/Edinburgh-Genome-Foundry/geneblocks",
keywords="DNA sequence blocks diff synthetic-biology bioinformatics",
- packages=find_packages(exclude='docs'),
- install_requires=['numpy', 'Biopython', 'dna_features_viewer',
- 'networkx', 'python-Levenshtein'])
+ packages=find_packages(exclude="docs"),
+ install_requires=[
+ "numpy",
+ "Biopython",
+ "dna_features_viewer",
+ "networkx",
+ "python-Levenshtein",
+ ],
+)
 -Licence
--------
+License = MIT
+-------------
Geneblocks is an open-source software originally written at the `Edinburgh Genome Foundry
-Licence
--------
+License = MIT
+-------------
Geneblocks is an open-source software originally written at the `Edinburgh Genome Foundry