A package for various forest mensuration and biometrics functions in Julia
ForestBiometrics.jl is a package for working with forest measurements data and growth and yield modeling. This package was inspired by its R counterpart, the lmfor package with the addition of equations from the western US. For more info on lmfor, please see http://cs.uef.fi/~lamehtat/rcodes.htm
It is my hope that the package not only ports existing functionality from other languages, but also showcases the abilities of Julia as a programming language and how concepts like multiple dispatch and metaprogramming can be used to solve domain-specific problems.
] add https://github.com/Crghilardi/ForestBiometrics.jl
If you are interested in this package and would like to contribute, feel free to submit an issue or pull request.
ForestBiometrics exports a Tree type and a Stand type for dispatch. The package was designed with fully expanded treelists in mind and makes no effort to accomodate plot data and plot compilation.
A Tree is a minimalistic container of fields that are absolutely needed for basic measurements. Future work will focus on the ability to add arbitrary fields as needed.
struct Tree
diameter::Real
height::Union{Real,Missing}
species::Union{AbstractString,Real}
expansion::Real
end
Likewise a Stand is mainly an Array of Tree objects with some very minimal summary info fields
mutable struct Stand
treelist::Array{Tree,1}
basal_area::Union{Real,Missing}
trees_per_area::Union{Real,Missing}
qmd::Union{Real,Missing}
end
using ForestBiometrics
using DelimitedFiles
using Plots
#read csv with tree data (upland HW stand from northeast USA)
datapath = joinpath(dirname(pathof(ForestBiometrics)), "..", "test/StandExam_data.csv")
data = readdlm(datapath,',',header=true)
tl = Tree[]
#read data and create array of Tree
for i in eachrow(data[1])
push!(tl,Tree(i[7], i[8], i[6], i[9]))
end
#create a Stand from the array of Tree
stand = Stand(tl)If just given a treelist, stand basal area, trees per area and qmd are intialized as missing. Once we have Trees and a Stand created we can calculate a number of summary information including:
#basal area of a single tree or the entire stand
basal_area(tl[1])
basal_area(stand)
#if the diameters are in cm
basal_area(tl[1],metric)
trees_per_area(stand)
qmd(stand)
sdi(stand)We can update the Stand object summary field by using the bang version (!) of the functions
basal_area!(stand)
trees_per_area!(stand)
qmd!(stand)
The package also exports a HeightModel that generalizes the process of estimating height from diameter using arbitrary model forms and parameters
#create Tree with diameter but no height
t1 = Tree(10.0,missing,"DF",1)
#make dictionary of species specific parameters
FVS_IE=Dict{String,Array{Float64}}(
"WP"=>[5.19988 -9.26718],
"WL"=>[4.97407 -6.78347],
"DF"=>[4.81519 -7.29306],
"GF"=>[5.00233 -8.19365])
#create HeightModel object using an named model form (exported by package) and the parameter dict above.
#See src/height.jl for full list of existing #model forms.
wyk = HeightModel(wyckoff,FVS_IE)
estimate_height(t1, wyk)
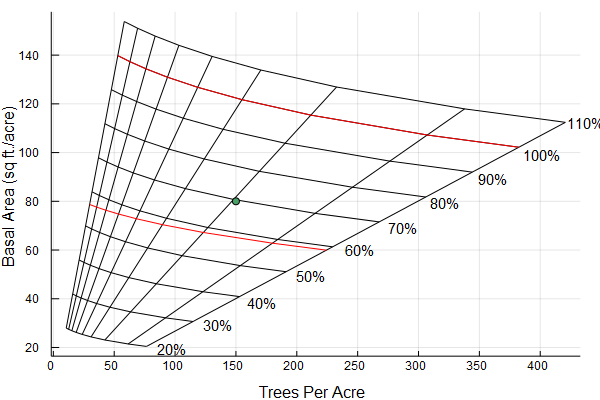
Gingrich stocking guides
gingrich_chart(stand)
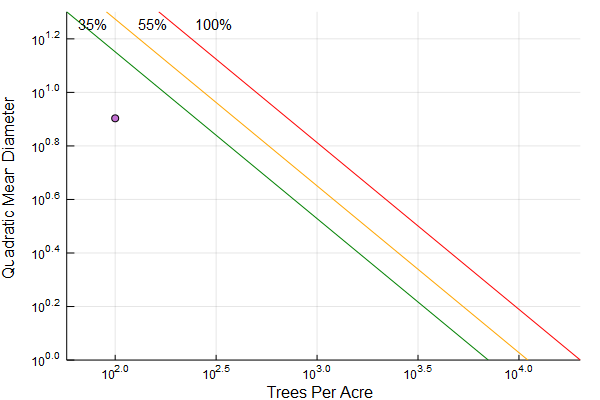
SDI chart with lines at 100%, 55% and 35% max SDI
reineke_chart(stand)
#or can define a max sdi explicity reineke_chart(stand; maxsdi = 250)